7S0L
 
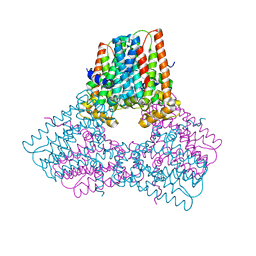 | |
7S0H
 
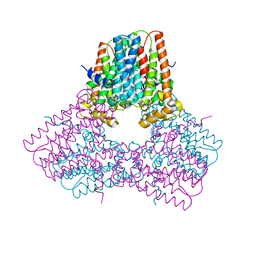 | |
7S0A
 
 | |
7S09
 
 | |
7S0M
 
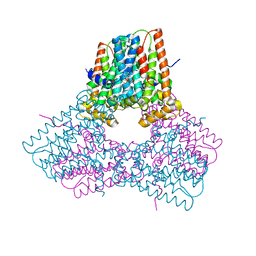 | |
3L27
 
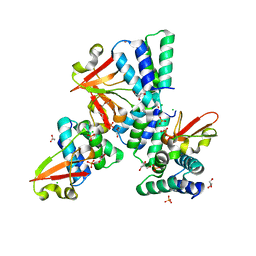 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain R312A mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7SI9
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with PF-07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-10-12 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
3L26
 
 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain bound to 8 bp dsRNA | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Polymerase cofactor VP35, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7SBB
 
 | |
7S9V
 
 | | DrmAB:ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DrmA, DrmB | | Authors: | Bravo, J.P.K, Taylor, D.W, Brouns, S.J.J, Aparicio-Maldonado, C. | | Deposit date: | 2021-09-21 | | Release date: | 2022-06-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for broad anti-phage immunity by DISARM.
Nat Commun, 13, 2022
|
|
7SBA
 
 | |
7S9W
 
 | | Structure of DrmAB:ADP:DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*T)-3'), DrmA, ... | | Authors: | Bravo, J.P.K, Taylor, D.W, Brounds, S.J.J, Aparicio-Maldonado, C. | | Deposit date: | 2021-09-21 | | Release date: | 2022-06-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for broad anti-phage immunity by DISARM.
Nat Commun, 13, 2022
|
|
3LW8
 
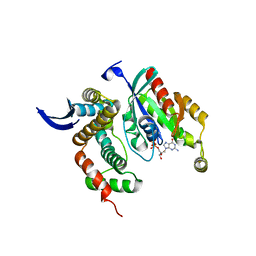 | | Shigella IpgB2 in complex with human RhoA, GDP and Mg2+ (complex A) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, IpgB2, MAGNESIUM ION, ... | | Authors: | Klink, B.U, Barden, S, Heidler, T.V, Borchers, C, Ladwein, M, Stradal, T.E.B, Rottner, K, Heinz, D.W. | | Deposit date: | 2010-02-23 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Shigella IPGB2 in complex with human RhoA: Implications for the mechanism of bacterial GEF-mimicry
J.Biol.Chem., 285, 2010
|
|
7SGJ
 
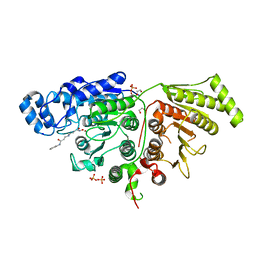 | |
7SGI
 
 | |
7SGG
 
 | |
7SGK
 
 | |
3LGK
 
 | | D99N Epi-isozizaene synthase | | Descriptor: | Epi-isozizaene synthase, SULFATE ION | | Authors: | Aaron, J.A, Lin, X, Cane, D.E, Christianson, D.W. | | Deposit date: | 2010-01-20 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structure of Epi-Isozizaene Synthase from Streptomyces coelicolor A3(2), a Platform for New Terpenoid Cyclization Templates
Biochemistry, 49, 2010
|
|
3LG5
 
 | | F198A Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, ... | | Authors: | Aaron, J.A, Lin, X, Cane, D.E, Christianson, D.W. | | Deposit date: | 2010-01-19 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structure of Epi-Isozizaene Synthase from Streptomyces coelicolor A3(2), a Platform for New Terpenoid Cyclization Templates
Biochemistry, 49, 2010
|
|
2LZH
 
 | | THE STRUCTURES OF THE MONOCLINIC AND ORTHORHOMBIC FORMS OF HEN EGG-WHITE LYSOZYME AT 6 ANGSTROMS RESOLUTION. | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Artymiuk, P.J, Blake, C.C.F, Rice, D.W, Wilson, K.S. | | Deposit date: | 1981-06-29 | | Release date: | 1981-09-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | The Structures of the Monoclinic and Orthorhombic Forms of Hen Egg-White Lysozyme at 6 Angstroms Resolution.
Acta Crystallogr.,Sect.B, 38, 1982
|
|
3LBX
 
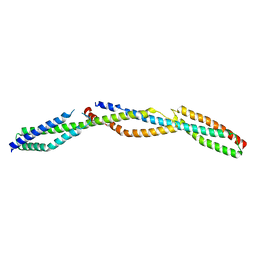 | | Crystal Structure of the Erythrocyte Spectrin Tetramerization Domain Complex | | Descriptor: | Spectrin alpha chain, erythrocyte, Spectrin beta chain | | Authors: | Ipsaro, J.J, Harper, S.L, Messick, T.E, Marmorstein, R, Mondragon, A, Speicher, D.W. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and functional interpretation of the erythrocyte spectrin tetramerization domain complex.
Blood, 115, 2010
|
|
3LP4
 
 | |
2N0N
 
 | | NMR solution structure for lactam (5,9) 11mer | | Descriptor: | lactam (5,9) 11mer peptide | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-10 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
3LX4
 
 | | Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA(deltaEFG) | | Descriptor: | ACETATE ION, CHLORIDE ION, Fe-hydrogenase, ... | | Authors: | Mulder, D.W, Boyd, E.S, Sarma, R, Lange, R.K, Endrizzi, J.A, Broderick, J.B, Peters, J.W. | | Deposit date: | 2010-02-24 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA(DeltaEFG).
Nature, 465, 2010
|
|
3LXR
 
 | | Shigella IpgB2 in complex with human RhoA and GDP (complex C) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, IpgB2, SULFATE ION, ... | | Authors: | Klink, B.U, Barden, S, Heidler, T.V, Borchers, C, Ladwein, M, Stradal, T.E.B, Rottner, K, Heinz, D.W. | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of Shigella IPGB2 in complex with human RhoA: Implications for the mechanism of bacterial GEF-mimicry
J.Biol.Chem., 285, 2010
|
|
