8H73
 
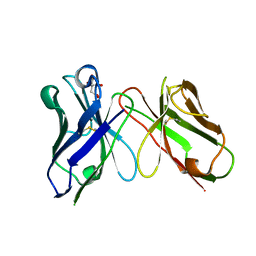 | | Crystal structure of antibody scFv against M2e Influenza peptide | | Descriptor: | CITRATE ANION, Single Chain Variable Fragment | | Authors: | Kumar, U, Madni, Z.K, Gaur, V, Salunke, D.M. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A structure and knowledge-based combinatorial approach to engineering universal scFv antibodies against influenza M2 protein.
J.Biomed.Sci., 30, 2023
|
|
8H3C
 
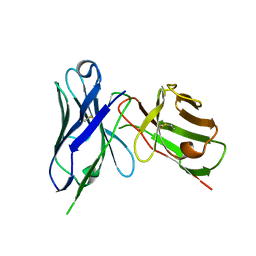 | | Crystal structure of M2e Influenza peptide in complex with antibody scFv | | Descriptor: | Matrix protein 2, Single Chain Variable Fragment | | Authors: | Kumar, U, Madni, Z.K, Gaur, V, Salunke, D.M. | | Deposit date: | 2022-10-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | A structure and knowledge-based combinatorial approach to engineering universal scFv antibodies against influenza M2 protein.
J.Biomed.Sci., 30, 2023
|
|
8H3B
 
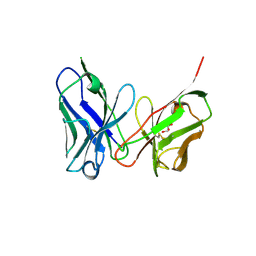 | | Crystal structure of antibody scFv against M2e Influenza peptide | | Descriptor: | GLYCEROL, Single Chain Variable Fragment | | Authors: | Kumar, U, Madni, Z.K, Gaur, V, Salunke, D.M. | | Deposit date: | 2022-10-08 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A structure and knowledge-based combinatorial approach to engineering universal scFv antibodies against influenza M2 protein.
J.Biomed.Sci., 30, 2023
|
|
7ANM
 
 | | Nudaurelia capensis omega virus capsid: virus-like particles expressed in Nicotiana benthamiana | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Ribeiro, J.R.S, Domitrovic, T, Hesketh, E.L, Scarff, C.A, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2020-10-12 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Plant-expressed virus-like particles reveal the intricate maturation process of a eukaryotic virus.
Commun Biol, 4, 2021
|
|
7ATA
 
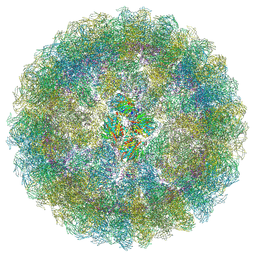 | | Nudaurelia capensis omega virus procapsid: virus-like particles expressed in Nicotiana benthamiana | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Ribeiro, J.R.S, Domitrovic, T, Hesketh, E.L, Scarff, C.A, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2020-10-29 | | Release date: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (6.63 Å) | | Cite: | Plant-expressed virus-like particles reveal the intricate maturation process of a eukaryotic virus.
Commun Biol, 4, 2021
|
|
7JN0
 
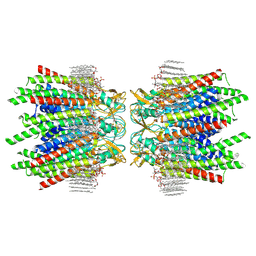 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 2 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.A, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-08-03 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JJP
 
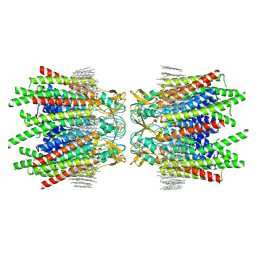 | | Sheep Connexin-50 at 1.9 angstroms resolution by CryoEM | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
4XIA
 
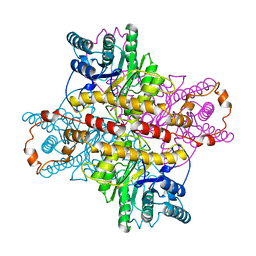 | | STRUCTURES OF D-XYLOSE ISOMERASE FROM ARTHROBACTER STRAIN B3728 CONTAINING THE INHIBITORS XYLITOL AND D-SORBITOL AT 2.5 ANGSTROMS AND 2.3 ANGSTROMS RESOLUTION, RESPECTIVELY | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, sorbitol | | Authors: | Henrick, K, Collyer, C.A, Blow, D.M. | | Deposit date: | 1989-07-05 | | Release date: | 1990-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of D-xylose isomerase from Arthrobacter strain B3728 containing the inhibitors xylitol and D-sorbitol at 2.5 A and 2.3 A resolution, respectively.
J.Mol.Biol., 208, 1989
|
|
7BT6
 
 | |
4YO4
 
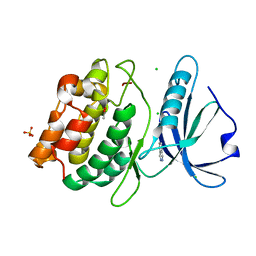 | | Crystal Structure of DAPK1 catalytic domain in complex with the hinge binding fragment phthalazine | | Descriptor: | ACETATE ION, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Grum-Tokars, V.L, Roy, S.M, Minasov, G, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2015-03-11 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of DAPK1 catalytic domain in complex with hinge binding fragments
To Be Published
|
|
7BTB
 
 | |
7JN1
 
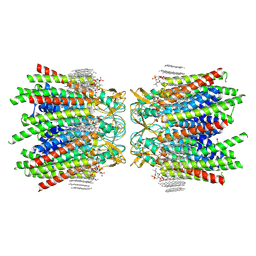 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-08-03 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7BNT
 
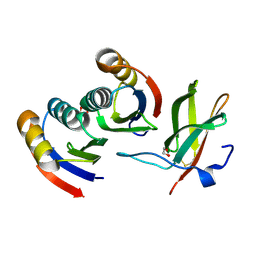 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikD with a predicted ancestral HMA domain of Pik-1 from Oryza spp. | | Descriptor: | 1,2-ETHANEDIOL, AVR-Pik protein, Predicted ancestral HMA domain of Pik-1 from Oryza spp. | | Authors: | Bialas, A, Langner, T, Harant, A, Contreras, M.P, Stevenson, C.E.M, Lawson, D.M, Sklenar, J, Kellner, R, Moscou, M.J, Terauchi, R, Banfield, M.J, Kamoun, S. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Two NLR immune receptors acquired high-affinity binding to a fungal effector through convergent evolution of their integrated domain.
Elife, 10, 2021
|
|
7BE6
 
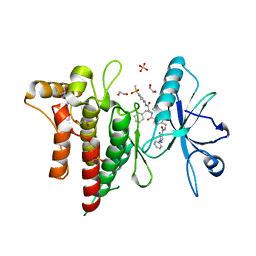 | | Structure of DDR1 receptor tyrosine kinase in complex with inhibitor SR159 | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-N-(4-(((2S)-4-cyclohexyl-1-((1-(methylsulfonyl)piperidin-3-yl)amino)-1-oxobutan-2-yl)carbamoyl)benzyl)-1-phenyl-1H-pyrazole-4-carboxamide, Epithelial discoidin domain-containing receptor 1, ... | | Authors: | Pinkas, D.M, Bufton, J.C, Roehm, S, Joerger, A.C, Knapp, S, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87081933 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
7JMD
 
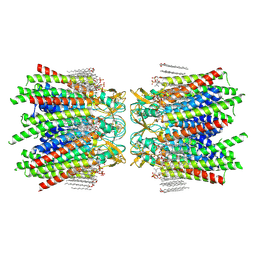 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 1 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JM9
 
 | | Sheep Connexin-50 at 2.5 angstroms reoslution, Lipid Class 2 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.A, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
8IA6
 
 | |
7JLW
 
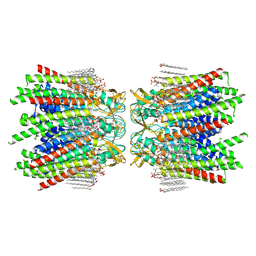 | | Sheep Connexin-50 at 2.5 angstroms resolution, Lipid Class 1 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7BM8
 
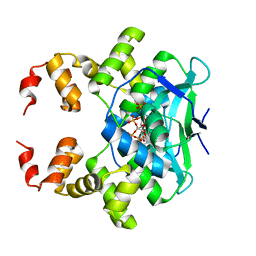 | | Crystal structure of the C-terminally truncated chromosome-partitioning protein ParB from Caulobacter crescentus complexed with CTP-gamma-S | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Chromosome-partitioning protein ParB, MAGNESIUM ION | | Authors: | Jalal, A.S, Tran, N.T, Stevenson, C.E.M, Lawson, D.M, Le, T.B.K. | | Deposit date: | 2021-01-19 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A CTP-dependent gating mechanism enables ParB spreading on DNA.
Elife, 10, 2021
|
|
7JMC
 
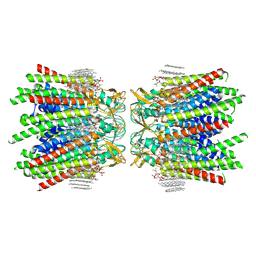 | | Sheep Connexin-50 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
3NG1
 
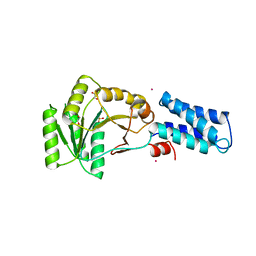 | | N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, SIGNAL SEQUENCE RECOGNITION PROTEIN FFH, ... | | Authors: | Freymann, D.M, Stroud, R.M, Walter, P. | | Deposit date: | 1998-09-13 | | Release date: | 1999-07-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional changes in the structure of the SRP GTPase on binding GDP and Mg2+GDP.
Nat.Struct.Biol., 6, 1999
|
|
8TXL
 
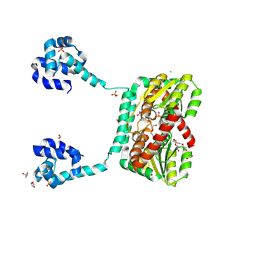 | |
8TX9
 
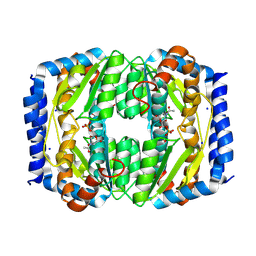 | |
8ULM
 
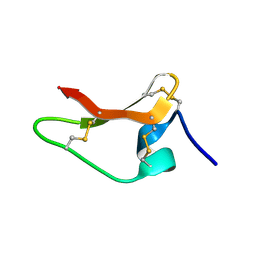 | |
8V1T
 
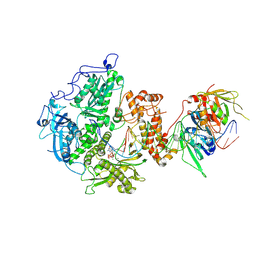 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and acyclovir triphosphate in closed conformation | | Descriptor: | ACYCLOVIR TRIPHOSPHATE, DNA polymerase, DNA polymerase processivity factor, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
