6TB7
 
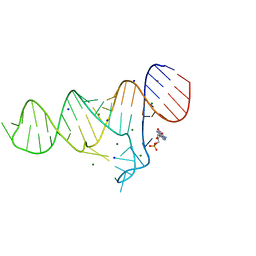 | |
6TF2
 
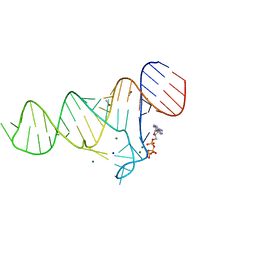 | |
6HCT
 
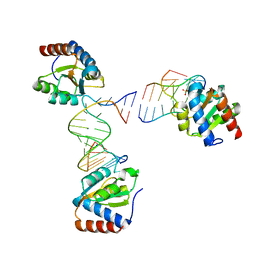 | |
6TFH
 
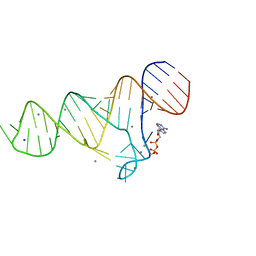 | |
6I5X
 
 | | Crystal structure of Aspergillus fumigatus phosphomannomutase | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Zhang, Y, Raimi, O.G, Ferenbach, A.T, van Aalten, D.M.F. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Aspergillus fumigatus phosphomannomutase
To Be Published
|
|
6IBO
 
 | | Catalytic deficiency of O-GlcNAc transferase leads to X-linked intellectual disability | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, ALA-VAL-SER-ARG-ALA, PHOSPHATE ION, ... | | Authors: | Gundogdu, M, van Aalten, D.M.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Catalytic deficiency of O-GlcNAc transferase leads to X-linked intellectual disability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4WPG
 
 | | Group A Streptococcus GacA is an essential dTDP-4-dehydrorhamnose reductase (RmlD) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, ... | | Authors: | van der Beek, S.L, Le Breton, Y, Ferenbach, A.T, van Aalten, D.M.F, Navratilova, I, McIver, K, van Sorge, N.M, Dorfmueller, H.C. | | Deposit date: | 2014-10-18 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | GacA is essential for Group A Streptococcus and defines a new class of monomeric dTDP-4-dehydrorhamnose reductases (RmlD).
Mol.Microbiol., 98, 2015
|
|
4XI9
 
 | | Human OGT in complex with UDP-5S-GlcNAc and substrate peptide (RBL2) | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, Retinoblastoma-like protein 2, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Schimpl, M, van Aalten, D.M.F. | | Deposit date: | 2015-01-06 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The active site of O-GlcNAc transferase imposes constraints on substrate sequence.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XLG
 
 | | C. glabrata Slx1 in complex with Slx4CCD. | | Descriptor: | CHLORIDE ION, Structure-specific endonuclease subunit SLX1, Structure-specific endonuclease subunit SLX4, ... | | Authors: | Gaur, V, Wyatt, H.D.M, Komorowska, W, Szczepanowski, R.H, de Sanctis, D, Gorecka, K.M, West, S.C, Nowotny, M. | | Deposit date: | 2015-01-13 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and Mechanistic Analysis of the Slx1-Slx4 Endonuclease.
Cell Rep, 10, 2015
|
|
4XIF
 
 | | Human OGT in complex with UDP-5S-GlcNAc and substrate peptide (keratin-7) | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, Keratin, type II cytoskeletal 7, ... | | Authors: | Schimpl, M, van Aalten, D.M.F. | | Deposit date: | 2015-01-06 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The active site of O-GlcNAc transferase imposes constraints on substrate sequence.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XM5
 
 | | C. glabrata Slx1. | | Descriptor: | CHLORIDE ION, Structure-specific endonuclease subunit SLX1, ZINC ION | | Authors: | Gaur, V, Wyatt, H.D.M, Komorowska, W, Szczepanowski, R.H, de Sanctis, D, Gorecka, K.M, West, S.C, Nowotny, M. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural and Mechanistic Analysis of the Slx1-Slx4 Endonuclease.
Cell Rep, 10, 2015
|
|
5CNQ
 
 | | Crystal structure of the Holliday junction-resolving enzyme GEN1 (WT) in complex with product DNA, Mg2+ and Mn2+ ions | | Descriptor: | DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MANGANESE (II) ION, Nuclease-like protein, ... | | Authors: | Liu, Y.J, Freeman, A.D.J, Declais, A.C, Wilson, T.J, Gartner, A, Lilley, D.M.J. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Crystal Structure of a Eukaryotic GEN1 Resolving Enzyme Bound to DNA.
Cell Rep, 13, 2015
|
|
5CO8
 
 | | Crystal structure of the Holliday junction-resolving enzyme GEN1 (WT) in complex with product DNA and Mg2+ ion | | Descriptor: | DNA (31-MER), DNA (5'-D(*AP*GP*AP*CP*TP*GP*CP*AP*GP*TP*TP*GP*AP*GP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*A)-3'), ... | | Authors: | Liu, Y.J, Freeman, A.D.J, Declais, A.C, Wilson, T.J, Gartner, A, Lilley, D.M.J. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a Eukaryotic GEN1 Resolving Enzyme Bound to DNA.
Cell Rep, 13, 2015
|
|
3LIM
 
 | | Crystal structure of the pore forming toxin frac from sea anemone actinia fragacea | | Descriptor: | Fragaceatoxin C, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Mechaly, A.E, Bellomio, A, Morante, K, Gonzalez-Manas, J.M, Guerin, D.M.A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the oligomerization and architecture of eukaryotic membrane pore-forming toxins.
Structure, 19, 2011
|
|
4ZXL
 
 | | CpOGA D298N in complex with Drosophila HCF -derived Thr-O-GlcNAc peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, NAG-PRO-SER-THR-ALA-Thr-O-GlcNAc containing peptide from drosophila HCF, ... | | Authors: | Selvan, N, van Aalten, D.M.F. | | Deposit date: | 2015-05-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A mutant O-GlcNAcase as a probe to reveal global dynamics of protein O-GlcNAcylation during Drosophila embryonic development.
Biochem.J., 470, 2015
|
|
6GRC
 
 | | eukaryotic junction-resolving enzyme GEN-1 binding with Sodium | | Descriptor: | DNA (5'-D(*TP*AP*CP*CP*CP*AP*CP*CP*AP*CP*CP*GP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Lilley, D.M.J, Liu, Y, Freeman, D.J. | | Deposit date: | 2018-06-11 | | Release date: | 2018-09-26 | | Last modified: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.
Nucleic Acids Res., 46, 2018
|
|
6GRD
 
 | | eukaryotic junction-resolving enzyme GEN-1 binding with Cesium | | Descriptor: | CESIUM ION, DNA (5'-D(*TP*AP*CP*CP*CP*AP*CP*CP*AP*CP*CP*GP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), ... | | Authors: | Lilley, D.M.J, Liu, Y, Freeman, D.J. | | Deposit date: | 2018-06-11 | | Release date: | 2018-09-26 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.
Nucleic Acids Res., 46, 2018
|
|
5DIY
 
 | | Thermobaculum terrenum O-GlcNAc hydrolase mutant - D120N | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hyaluronidase, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 | | Authors: | Ostrowski, A, Gundogdu, M, Ferenbach, A.T, Lebedev, A, van Aalten, D.M.F. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Evidence for a Functional O-Linked N-Acetylglucosamine (O-GlcNAc) System in the Thermophilic Bacterium Thermobaculum terrenum.
J.Biol.Chem., 290, 2015
|
|
5A01
 
 | | O-GlcNAc transferase from Drososphila melanogaster | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, O-GLYCOSYLTRANSFERASE | | Authors: | Mariappa, D, Zheng, X, Schimpl, M, Raimi, O, Rafie, K, Ferenbach, A.T, Mueller, H.J, van Aalten, D.M.F. | | Deposit date: | 2015-04-15 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Dual functionality of O-GlcNAc transferase is required for Drosophila development.
Open Biol, 5, 2015
|
|
6GRB
 
 | | eukaryotic junction-resolving enzyme GEN-1 binding with Potassium | | Descriptor: | DNA (5'-D(*TP*AP*CP*CP*CP*AP*CP*CP*AP*CP*CP*GP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Lilley, D.M.J, Liu, Y, Freeman, D.J. | | Deposit date: | 2018-06-11 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.
Nucleic Acids Res., 46, 2018
|
|
3ICB
 
 | | THE REFINED STRUCTURE OF VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN FROM BOVINE INTESTINE. MOLECULAR DETAILS, ION BINDING, AND IMPLICATIONS FOR THE STRUCTURE OF OTHER CALCIUM-BINDING PROTEINS | | Descriptor: | CALCIUM ION, CALCIUM-BINDING PROTEIN, SULFATE ION | | Authors: | Szebenyi, D.M.E, Moffat, K. | | Deposit date: | 1986-09-09 | | Release date: | 1986-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The refined structure of vitamin D-dependent calcium-binding protein from bovine intestine. Molecular details, ion binding, and implications for the structure of other calcium-binding proteins.
J.Biol.Chem., 261, 1986
|
|
6FZ0
 
 | |
5BNW
 
 | | The active site of O-GlcNAc transferase imposes constraints on substrate sequence | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, laminB1 residues 179-191 | | Authors: | Pathak, S, Alonso, J, Schimpl, M, Rafie, K, Blair, D.E, Borodkin, V.S, Albarbarawi, O, van Aalten, D.M.F. | | Deposit date: | 2015-05-26 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The active site of O-GlcNAc transferase imposes constraints on substrate sequence.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5DJS
 
 | | Thermobaculum terrenum O-GlcNAc transferase mutant - K341M | | Descriptor: | Tetratricopeptide TPR_2 repeat protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Ostrowski, A, Gundogdu, M, Ferenbach, A.T, Lebedev, A, van Aalten, D.M.F. | | Deposit date: | 2015-09-02 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for a Functional O-Linked N-Acetylglucosamine (O-GlcNAc) System in the Thermophilic Bacterium Thermobaculum terrenum.
J.Biol.Chem., 290, 2015
|
|
3GNI
 
 | | Structure of STRAD and MO25 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, Protein Mo25, ... | | Authors: | Zeqiraj, E, Goldie, S, Alessi, D.R, van Aalten, D.M.F. | | Deposit date: | 2009-03-17 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | ATP and MO25alpha regulate the conformational state of the STRADalpha pseudokinase and activation of the LKB1 tumour suppressor
Plos Biol., 7, 2009
|
|
