3GU4
 
 | |
3GU7
 
 | |
3GU6
 
 | |
3GUB
 
 | | Crystal structure of DAPKL93G complexed with N6-(2-Phenylethyl)adenosine | | Descriptor: | 9-alpha-L-lyxofuranosyl-N-(2-phenylethyl)-9H-purin-6-amine, Death-associated protein kinase 1, SULFATE ION | | Authors: | McNamara, L.K, Schumacher, A.M, Schavocky, J.S, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2009-03-29 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structures of the DAPK gatekeeper mutant complexed with N6-modified adenosine analogs.
To be Published
|
|
3GU8
 
 | | Crystal structure of DAPKL93G with N6-cyclopentyladenosine | | Descriptor: | Death-associated protein kinase 1, N6-cyclopentyladenosine | | Authors: | McNamara, L.K, Schumacher, A.M, Schavocky, J.S, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2009-03-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the DAPK gatekeeper mutant with N6-modified adenosine analogs.
To be Published
|
|
3HHN
 
 | | Crystal structure of class I ligase ribozyme self-ligation product, in complex with U1A RBD | | Descriptor: | Class I ligase ribozyme, self-ligation product, MAGNESIUM ION, ... | | Authors: | Shechner, D.M, Grant, R.A, Bagby, S.C, Bartel, D.P. | | Deposit date: | 2009-05-15 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.987 Å) | | Cite: | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
3HPL
 
 | | KcsA E71H-F103A mutant in the closed state | | Descriptor: | Antibody Fab heavy chain, Antibody fab light chain, POTASSIUM ION, ... | | Authors: | Cuello, L.G, Jogini, V, Cortes, D.M, Perozo, E. | | Deposit date: | 2009-06-04 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the coupling between activation and inactivation gates in K(+) channels.
Nature, 466, 2010
|
|
3I5L
 
 | | Allosteric Modulation of DNA by Small Molecules | | Descriptor: | (22R,51R)-22,51-diamino-5,11,17,28,34,40,46,57-octamethyl-2,5,8,11,14,17,20,25,28,31,34,37,40,43,46,49,54,57,60,61,64,6 5-docosaazanonacyclo[54.2.1.1~4,7~.1~10,13~.1~16,19~.1~27,30~.1~33,36~.1~39,42~.1~45,48~]hexahexaconta-1(58),4(66),6,10( 65),12,16(64),18,27(63),29,33(62),35,39(61),41,45(60),47,56(59)-hexadecaene-3,9,15,21,26,32,38,44,50,55-decone, 5'-D(*CP*CP*AP*GP*GP*(C38)P*CP*TP*GP*G)-3', CALCIUM ION | | Authors: | Chenoweth, D.M, Dervan, P.B. | | Deposit date: | 2009-07-05 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Allosteric modulation of DNA by small molecules
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ILQ
 
 | | Structure of mCD1d with bound glycolipid BbGL-2c from Borrelia burgdorferi | | Descriptor: | (2S)-3-(alpha-D-galactopyranosyloxy)-2-(hexadecanoyloxy)propyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M. | | Deposit date: | 2009-08-07 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3INU
 
 | | Crystal structure of an unbound KZ52 neutralizing anti-Ebolavirus antibody. | | Descriptor: | GLYCEROL, KZ52 antibody fragment heavy chain, KZ52 antibody fragment light chain, ... | | Authors: | Lee, J.E, Fusco, M.L, Abelson, D.M, Hessell, A.J, Burton, D.R, Saphire, E.O. | | Deposit date: | 2009-08-12 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Techniques and tactics used in determining the structure of the trimeric ebolavirus glycoprotein.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3ILP
 
 | | Structure of mCD1d with bound glycolipid BbGL-2f from Borrelia burgdorferi | | Descriptor: | (2S)-3-(alpha-D-galactopyranosyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl (9Z,12Z)-octadeca-9,12-dienoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M. | | Deposit date: | 2009-08-07 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IC1
 
 | | Crystal structure of zinc-bound succinyl-diaminopimelate desuccinylase from Haemophilus influenzae | | Descriptor: | GLYCEROL, SULFATE ION, Succinyl-diaminopimelate desuccinylase, ... | | Authors: | Nocek, B.P, Gillner, D.M, Holz, R.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for catalysis by the mono- and dimetalated forms of the dapE-encoded N-succinyl-L,L-diaminopimelic acid desuccinylase.
J.Mol.Biol., 397, 2010
|
|
3I5E
 
 | |
3I5F
 
 | | Crystal structure of squid MG.ADP myosin S1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin catalytic light chain LC-1, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
3ISZ
 
 | | Crystal structure of mono-zinc form of succinyl-diaminopimelate desuccinylase from Haemophilus influenzae | | Descriptor: | SULFATE ION, Succinyl-diaminopimelate desuccinylase, ZINC ION | | Authors: | Nocek, B.P, Gillner, D.M, Holz, R.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for catalysis by the mono- and dimetalated forms of the dapE-encoded N-succinyl-L,L-diaminopimelic acid desuccinylase.
J.Mol.Biol., 397, 2010
|
|
3I2M
 
 | | The Crystal Structure of PF-8, the DNA Polymerase Accessory Subunit from Kaposi s Sarcoma-Associated Herpesvirus | | Descriptor: | ORF59 | | Authors: | Baltz, J.L, Filman, D.J, Ciustea, M, Silverman, J.E.Y, Lautenschlager, C.L, Coen, D.M, Ricciardi, R.P, Hogle, J.M. | | Deposit date: | 2009-06-29 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The crystal structure of PF-8, the DNA polymerase accessory subunit from Kaposi's sarcoma-associated herpesvirus.
J.Virol., 83, 2009
|
|
3I5G
 
 | | Crystal structure of rigor-like squid myosin S1 | | Descriptor: | CALCIUM ION, MALONATE ION, Myosin catalytic light chain LC-1, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
3HS7
 
 | | X-ray crystal structure of docosahexaenoic acid bound to the cyclooxygenase channel of cyclooxygenase-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchio, A.J, Simmons, D.M, Malkowski, M.G. | | Deposit date: | 2009-06-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of fatty acid substrate binding to cyclooxygenase-2.
J.Biol.Chem., 285, 2010
|
|
3I5I
 
 | | The crystal structure of squid myosin S1 in the presence of SO4 2- | | Descriptor: | CALCIUM ION, Myosin catalytic light chain LC-1, mantle muscle, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
3J05
 
 | | Three-dimensional structure of Dengue virus serotype 1 complexed with HMAb 14c10 Fab | | Descriptor: | envelope protein | | Authors: | Teoh, E.P, Kukkaro, P, Teo, E.W, Lim, A, Tan, T.T, Shi, P.Y, Yip, A, Schul, W, Leo, Y.S, Chan, S.H, Smith, K.G.C, Ooi, E.E, Kemeny, D.M, Ng, G, Ng, M.L, Alonso, S, Fisher, D, Hanson, B, Lok, S.M, MacAry, P.A. | | Deposit date: | 2011-04-01 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | The structural basis for serotype-specific neutralization of dengue virus by a human antibody.
Sci Transl Med, 4, 2012
|
|
3IYS
 
 | | Homology model of avian polyomavirus asymmetric unit | | Descriptor: | Major capsid protein VP1 | | Authors: | Shen, P.S, Enderlein, D, Nelson, C.D.S, Carter, W.S, Kawano, M, Xing, L, Swenson, R.D, Olson, N.H, Baker, T.S, Cheng, R.H, Atwood, W.J, Johne, R, Belnap, D.M. | | Deposit date: | 2010-04-19 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | The structure of avian polyomavirus reveals variably sized capsids, non-conserved inter-capsomere interactions, and a possible location of the minor capsid protein VP4.
Virology, 411, 2011
|
|
3J1A
 
 | | HK97-like fold fitted into 3D reconstruction of bacteriophage CW02 | | Descriptor: | capsid protein | | Authors: | Shen, P.S, Domek, M.J, Sanz-Garcia, E, Makaju, A, Taylor, R, Culumber, M, Breakwell, D.P, Prince, J.T, Belnap, D.M. | | Deposit date: | 2012-01-31 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Sequence and Structural Characterization of Great Salt Lake Bacteriophage CW02, a Member of the T7-Like Supergroup.
J.Virol., 86, 2012
|
|
3J1S
 
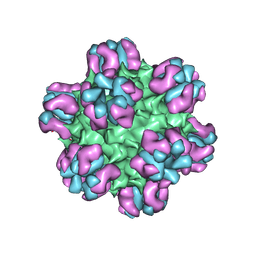 | |
3J3P
 
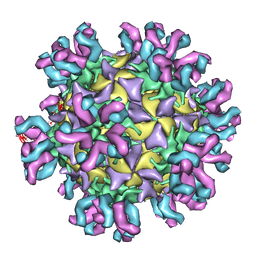 | | Conformational Shift of a Major Poliovirus Antigen Confirmed by Immuno-Cryogenic Electron Microscopy: 135S Poliovirus and C3-Fab Complex | | Descriptor: | C3 antibody, heavy chain, light chain, ... | | Authors: | Lin, J, Cheng, N, Hogle, J.M, Steven, A.C, Belnap, D.M. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Conformational shift of a major poliovirus antigen confirmed by immuno-cryogenic electron microscopy.
J.Immunol., 191, 2013
|
|
3J41
 
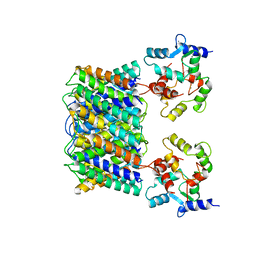 | | Pseudo-atomic model of the Aquaporin-0/Calmodulin complex derived from electron microscopy | | Descriptor: | CALCIUM ION, Calmodulin, Lens fiber major intrinsic protein | | Authors: | Reichow, S.L, Clemens, D.M, Freites, J.A, Nemeth-Cahalan, K.L, Heyden, M, Tobias, D.J, Hall, J.E, Gonen, T. | | Deposit date: | 2013-05-31 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Allosteric mechanism of water-channel gating by Ca(2+)-calmodulin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
