4BOJ
 
 | |
4CAV
 
 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a benzofuran ligand R0-09-4879 | | Descriptor: | 3-[[3-methyl-2-[[2,3,4-tris(fluoranyl)phenoxy]methyl]-1-benzofuran-4-yl]oxy]-N-(pyridin-3-ylmethyl)propan-1-amine, CHLORIDE ION, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Robinson, D.A, Fang, W, Raimi, O.G, Blair, D.E, Harrison, J, Ruda, G.F, Lockhart, D.E.A, Torrie, L.S, Wyatt, P.G, Gilbert, I.H, Van Aalten, D.M.F. | | Deposit date: | 2013-10-09 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | N-Myristoyltransferase is a Cell Wall Target in Aspergillus Fumigatus.
Acs Chem.Biol., 10, 2015
|
|
4C40
 
 | |
4C4W
 
 | | Structure of a rare, non-standard sequence k-turn bound by L7Ae protein | | Descriptor: | 50S RIBOSOMAL PROTEIN L7AE, DIHYDROGENPHOSPHATE ION, TSKT-23, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2013-09-09 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of a Rare, Non-Standard Sequence K-Turn Bound by L7Ae Protein
Nucleic Acids Res., 42, 2014
|
|
4BW0
 
 | |
4CDR
 
 | | Human O-GlcNAc transferase in complex with a bisubstrate inhibitor, Goblin1 | | Descriptor: | GOBLIN1, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE--PEPTIDE N-ACETYLGLUCOSAMINYLTRANSFERASE 110 KDA SUBUNIT, ... | | Authors: | Schimpl, M, Gundogdu, M, van Aalten, D.M.F. | | Deposit date: | 2013-11-05 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Bisubstrate Udp-Peptide Conjugates as Human O-Glcnac Transferase Inhibitors.
Biochem.J., 457, 2014
|
|
4B5R
 
 | | SAM-I riboswitch bearing the H. marismortui K-t-7 | | Descriptor: | BARIUM ION, POTASSIUM ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Daldrop, P, Lilley, D.M.J. | | Deposit date: | 2012-08-07 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The Plasticity of a Structural Motif in RNA: Structural Polymorphism of a Kink Turn as a Function of its Environment.
RNA, 19, 2013
|
|
4CAX
 
 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a pyrazole sulphonamide ligand (DDD85646) | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Raimi, O.G, Robinson, D.A, Fang, W, Blair, D.E, Harrison, J, Ruda, G.F, Lockhart, D.E.A, Torrie, L.S, Wyatt, P.G, Gilbert, I.H, Van Aalten, D.M.F. | | Deposit date: | 2013-10-09 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N-Myristoyltransferase is a Cell Wall Target in Aspergillus Fumigatus.
Acs Chem.Biol., 10, 2015
|
|
4AOB
 
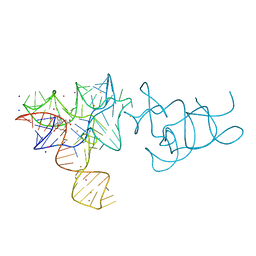 | | SAM-I riboswitch containing the T. solenopsae Kt-23 in complex with S- adenosyl methionine | | Descriptor: | BARIUM ION, POTASSIUM ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Schroeder, K.T, Daldrop, P, McPhee, S.A, Lilley, D.M.J. | | Deposit date: | 2012-03-25 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure and Folding of a Rare, Natural Kink Turn in RNA with an Aa Pair at the 2B2N Position.
RNA, 18, 2012
|
|
4BMA
 
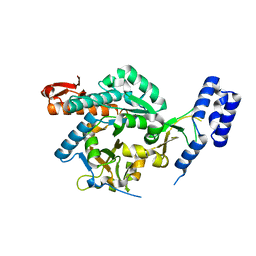 | | structural of Aspergillus fumigatus UDP-N-acetylglucosamine pyrophosphorylase | | Descriptor: | GLYCEROL, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE | | Authors: | Fang, W, Raimi, O.G, HurtadoGuerrero, R, vanAalten, D.M.F. | | Deposit date: | 2013-05-07 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Genetic and Structural Validation of Aspergillus Fumigatus Udp-N-Acetylglucosamine Pyrophosphorylase as an Antifungal Target.
Mol.Microbiol., 89, 2013
|
|
4BHU
 
 | | Crystal structure of BslA - A bacterial hydrophobin | | Descriptor: | CHLORIDE ION, GLYCEROL, UNCHARACTERIZED PROTEIN YUAB | | Authors: | Rao, F.V, Hobley, L, Ostrowski, A, Bromley, K.M, Porter, M, Prescott, A.R, Swedlow, J.R, MacPhee, C.E, van Aalten, D.M.F, Stanley-Wall, N.R. | | Deposit date: | 2013-04-08 | | Release date: | 2013-08-14 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Bsla is a Self-Assembling Bacterial Hydrophobin that Coats the Bacillus Subtilis Biofilm.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AG7
 
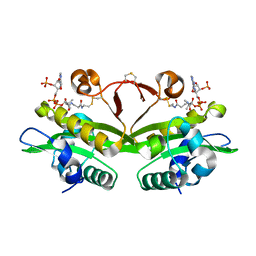 | | C. elegans glucosamine-6-phosphate N-acetyltransferase (GNA1): coenzyme A adduct | | Descriptor: | COENZYME A, GLUCOSAMINE-6-PHOSPHATE N-ACETYLTRANSFERASE | | Authors: | Dorfmueller, H.C, Fang, W, Rao, F.V, Blair, D.E, Attrill, H, Shepherd, S.M, van Aalten, D.M.F. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Biochemical Characterization of a Trapped Coenzyme a Adduct of Caenorhabditis Elegans Glucosamine-6-Phosphate N-Acetyltransferase 1.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4B7M
 
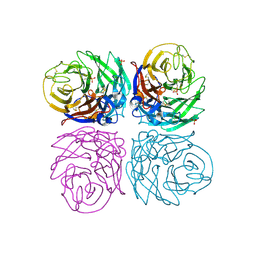 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | van der Vries, E, Vachieri, S.G, Xiong, X, Liu, J, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
4B7J
 
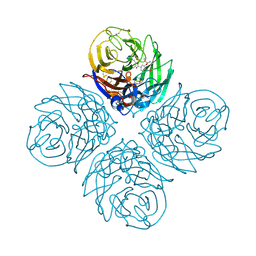 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | van der Vries, E, Vachieri, S.G, Xiong, X, Liu, J, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-20 | | Release date: | 2012-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
4B9D
 
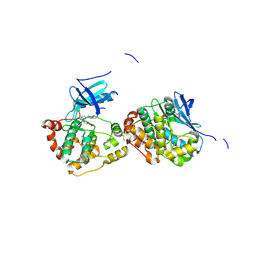 | | Crystal Structure of Human NIMA-related Kinase 1 (NEK1) with inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK1, ... | | Authors: | Elkins, J.M, Hanchuk, T.D.M, Lovato, D.V, Basei, F.L, Meirelles, G.V, Kobarg, J, Szklarz, M, Vollmar, M, Mahajan, P, Rellos, P, Zhang, Y, Krojer, T, Pike, A.C.W, Canning, P, von Delft, F, Raynor, J, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-09-04 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin.
Sci Rep, 7, 2017
|
|
4AG9
 
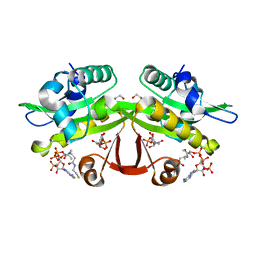 | | C. elegans glucosamine-6-phosphate N-acetyltransferase (GNA1): ternary complex with coenzyme A and GlcNAc | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, COENZYME A, ... | | Authors: | Dorfmueller, H.C, Fang, W, Rao, F.V, Blair, D.E, Attrill, H, Shepherd, S.M, van Aalten, D.M.F. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and Biochemical Characterization of a Trapped Coenzyme a Adduct of Caenorhabditis Elegans Glucosamine-6-Phosphate N-Acetyltransferase 1.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4APC
 
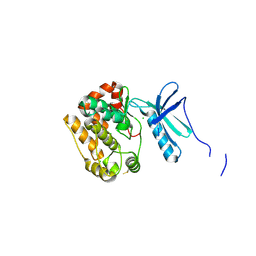 | | Crystal Structure of Human NIMA-related Kinase 1 (NEK1) | | Descriptor: | CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK1 | | Authors: | Elkins, J.M, Hanchuk, T.D.M, Lovato, D.V, Basei, F.L, Meirelles, G.V, Kobarg, J, Szklarz, M, Vollmar, M, Mahajan, P, Rellos, P, Zhang, Y, Krojer, T, Pike, A.C.W, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-04-02 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin.
Sci Rep, 7, 2017
|
|
4AY1
 
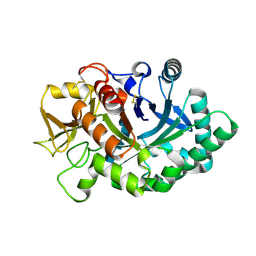 | | Human YKL-39 is a pseudo-chitinase with retained chitooligosaccharide binding properties | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE-3-LIKE PROTEIN 2 | | Authors: | Schimpl, M, Rush, C.L, Betou, M, Eggleston, I.M, Penman, G.A, Recklies, A.D, van Aalten, D.M.F. | | Deposit date: | 2012-06-17 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human Ykl-39 is a Pseudo-Chitinase with Retained Chitooligosaccharide-Binding Properties.
Biochem.J., 446, 2012
|
|
4AY5
 
 | | Human O-GlcNAc transferase (OGT) in complex with UDP and glycopeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GTAB1TIDE, UDP-N-ACETYLGLUCOSAMINE--PEPTIDE N-ACETYLGLUCOSAMINYL TRANSFERASE 110 KDA SUBUNIT, ... | | Authors: | Schimpl, M, Zheng, X, Blair, D.E, Schuettelkopf, A.W, Navratilova, I, Aristotelous, T, Ferenbach, A.T, Macnaughtan, M.A, Borodkin, V.S, van Aalten, D.M.F. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | O-Glcnac Transferase Invokes Nucleotide Sugar Pyrophosphate Participation in Catalysis
Nat.Chem.Biol., 8, 2012
|
|
4B7N
 
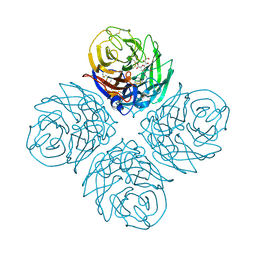 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE, ... | | Authors: | van der Vries, E, Vachieri, S.G, Xiong, X, Liu, J, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
4B7Q
 
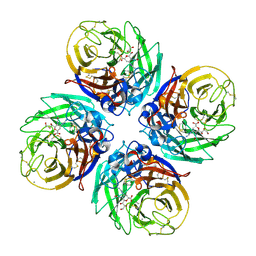 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE, ... | | Authors: | Liu, J, van der Vries, E, Vachieri, S.G, Xiong, X, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.728 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
4CS1
 
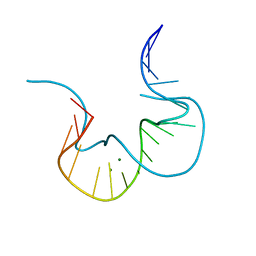 | |
4BQH
 
 | | Crystal structure of the uridine diphosphate N-acetylglucosamine pyrophosphorylase from Trypanosoma brucei in complex with inhibitor | | Descriptor: | (3S)-3-[2-(1,3-benzodioxol-5-yl)-2-oxidanylidene-ethyl]-4-bromanyl-5-methyl-3-oxidanyl-1H-indol-2-one, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE | | Authors: | Fang, W, Raimi, O.G, vanAalten, D.M.F. | | Deposit date: | 2013-05-30 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Novel Allosteric Inhibitor of the Uridine Diphosphate N-Acetylglucosamine Pyrophosphorylase from Trypanosoma Brucei.
Acs Chem.Biol., 8, 2013
|
|
4BWO
 
 | | The FedF adhesin from entrrotoxigenic Escherichia coli is a sulfate- binding lectin | | Descriptor: | BROMIDE ION, F18 FIMBRIAL ADHESIN AC, SULFATE ION | | Authors: | Lonardi, E, Moonens, K, Buts, L, de Boer, A.R, Olsson, J.D.M, Weiss, M.S, Fabre, E, Guerardel, Y, Deelder, A.M, Oscarson, S, Wuhrer, M, Bouckaert, J. | | Deposit date: | 2013-07-03 | | Release date: | 2013-08-28 | | Last modified: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia Coli
Biology, 2, 2013
|
|
4B7R
 
 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, J, van der Vries, E, Vachieri, S.G, Xiong, X, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
