1E6Z
 
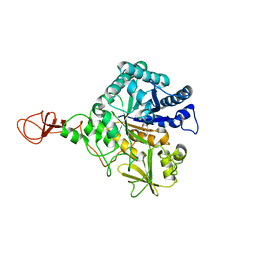 | | CHITINASE B FROM SERRATIA MARCESCENS WILDTYPE IN COMPLEX WITH CATALYTIC INTERMEDIATE | | Descriptor: | 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Komander, D, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2000-08-23 | | Release date: | 2001-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of a Family 18 Exo-Chitinase
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1NKO
 
 | | Energetic and structural basis of sialylated oligosaccharide recognition by the natural killer cell inhibitory receptor p75/AIRM1 or Siglec-7 | | Descriptor: | Sialic acid binding Ig-like lectin 7 | | Authors: | Dimasi, N, Attril, H, van Aalten, D.M.F, Moretta, L, Biassoni, R, Mariuzza, R.A. | | Deposit date: | 2003-01-03 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the saccharide-binding domain of the human natural killer cell inhibitory receptor p75/AIRM1.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OK9
 
 | | Decay accelerating factor (CD55): The structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, CHLORIDE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, R.A, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-21 | | Release date: | 2004-01-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1W1D
 
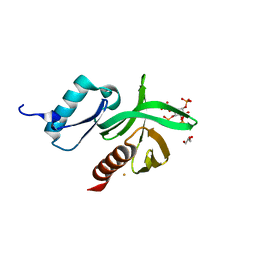 | | Crystal Structure of the PDK1 Pleckstrin Homology (PH) domain bound to Inositol (1,3,4,5)-tetrakisphosphate | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, GLYCEROL, GOLD ION, ... | | Authors: | Komander, D, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2004-06-21 | | Release date: | 2004-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights Into the Regulation of Pdk1 by Phosphoinositides and Inositol Phosphates
Embo J., 23, 2004
|
|
1W1H
 
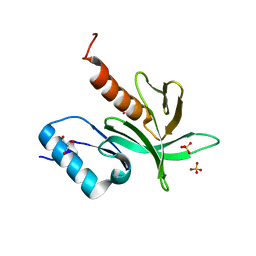 | | Crystal Structure of the PDK1 Pleckstrin Homology (PH) domain | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, GLYCEROL, SULFATE ION | | Authors: | Komander, D, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2004-06-21 | | Release date: | 2004-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Insights Into the Regulation of Pdk1 by Phosphoinositides and Inositol Phosphates
Embo J., 23, 2004
|
|
1H1W
 
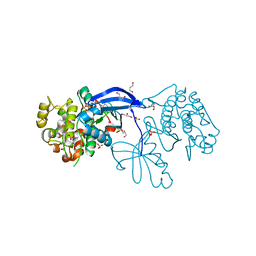 | | High resolution crystal structure of the human PDK1 catalytic domain | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Biondi, R.M, Komander, D, Thomas, C.C, Lizcano, J.M, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2002-07-23 | | Release date: | 2003-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Crystal Structure of the Human Pdk1 Catalytic Domain Defines the Regulatory Phosphopeptide Docking Site
Embo J., 21, 2003
|
|
1W1B
 
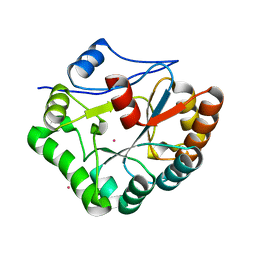 | |
1HBK
 
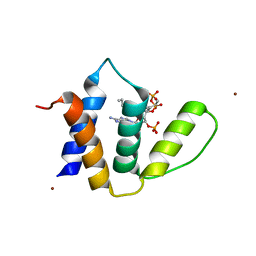 | | Acyl-CoA binding protein from Plasmodium falciparum | | Descriptor: | ACYL-COA BINDING PROTEIN, COENZYME A, MYRISTIC ACID, ... | | Authors: | van Aalten, D.M.F. | | Deposit date: | 2001-04-16 | | Release date: | 2001-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding site differences revealed by crystal structures of Plasmodium falciparum and bovine acyl-CoA binding protein.
J. Mol. Biol., 309, 2001
|
|
244D
 
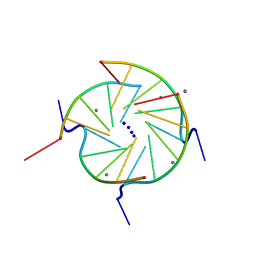 | | THE HIGH-RESOLUTION CRYSTAL STRUCTURE OF A PARALLEL-STRANDED GUANINE TETRAPLEX | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3'), SODIUM ION | | Authors: | Laughlan, G, Murchie, A.I.H, Norman, D.G, Moore, M.H, Moody, P.C.E, Lilley, D.M.J, Luisi, B. | | Deposit date: | 1995-10-19 | | Release date: | 1996-02-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The high-resolution crystal structure of a parallel-stranded guanine tetraplex.
Science, 265, 1994
|
|
2A3A
 
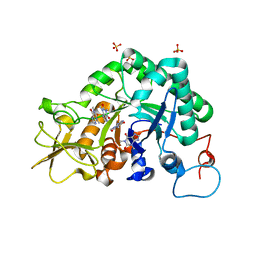 | | Crystal structure of Aspergillus fumigatus chitinase B1 in complex with theophylline | | Descriptor: | SULFATE ION, THEOPHYLLINE, chitinase | | Authors: | Rao, F.V, Andersen, O.A, Vora, K.A, DeMartino, J.A, van Aalten, D.M.F. | | Deposit date: | 2005-06-24 | | Release date: | 2005-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methylxanthine drugs are chitinase inhibitors: investigation of inhibition and binding modes.
Chem.Biol., 12, 2005
|
|
1H0I
 
 | | Complex of a chitinase with the natural product cyclopentapeptide argifin from Gliocladium | | Descriptor: | ARGIFIN, CHITINASE B, GLYCEROL, ... | | Authors: | Houston, D.R, Shiomi, K, Arai, N, Omura, S, Peter, M.G, Turberg, A, Synstad, B, Eijsink, V.G.H, Aalten, D.M.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Inhibited Complexes of a Chitinase with Natural Product Cyclopentapeptides - Peptide Mimicry of a Carbohydrate Substrate
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1H10
 
 | | HIGH RESOLUTION STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN OF PROTEIN KINASE B/AKT BOUND TO INS(1,3,4,5)-TETRAKISPHOPHATE | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, RAC-ALPHA SERINE/THREONINE KINASE | | Authors: | Thomas, C.C, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2002-07-01 | | Release date: | 2003-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High Resolution Structure of the Pleckstrin Homology Domain of Protein Kinase B/Akt Bound to Phosphatidylinositol (3,4,5)-Trisphosphate
Curr.Biol., 12, 2002
|
|
1R16
 
 | | Aplysia ADP ribosyl cyclase with bound pyridylcarbinol and R5P | | Descriptor: | 3-PYRIDINYLCARBINOL, ADP-ribosyl cyclase, ANY 5'-MONOPHOSPHATE NUCLEOTIDE | | Authors: | Love, M.L, Szebenyi, D.M.E, Kriksunov, I.A, Thiel, D.J, Munshi, C, Graeff, R, Lee, H.C, Hao, Q. | | Deposit date: | 2003-09-23 | | Release date: | 2004-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ADP-ribosyl cyclase; crystal structures reveal a covalent intermediate.
Structure, 12, 2004
|
|
2UY5
 
 | | ScCTS1_kinetin crystal structure | | Descriptor: | ENDOCHITINASE, N-(FURAN-2-YLMETHYL)-7H-PURIN-6-AMINE | | Authors: | Hurtado-Guerrero, R, van Aalten, D.M.F. | | Deposit date: | 2007-04-02 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Saccharomyces Cerevisiae Chitinase 1 and Screening-Based Discovery of Potent Inhibitors.
Chem.Biol., 14, 2007
|
|
2UY2
 
 | | ScCTS1_apo crystal structure | | Descriptor: | ENDOCHITINASE, GLYCEROL | | Authors: | Hurtado-Guerrero, R, Van Aalten, D.M.F. | | Deposit date: | 2007-04-02 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Saccharomyces Cerevisiae Chitinase 1 and Screening-Based Discovery of Potent Inhibitors.
Chem.Biol., 14, 2007
|
|
2UY4
 
 | | ScCTS1_acetazolamide crystal structure | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, ENDOCHITINASE | | Authors: | Hurtado-Guerrero, R, van Aalten, D.M.F. | | Deposit date: | 2007-04-02 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of Saccharomyces Cerevisiae Chitinase 1 and Screening-Based Discovery of Potent Inhibitors.
Chem.Biol., 14, 2007
|
|
2VXK
 
 | | Structural comparison between Aspergillus fumigatus and human GNA1 | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, COENZYME A, GLUCOSAMINE 6-PHOSPHATE ACETYLTRANSFERASE, ... | | Authors: | Hurtado-Guerrero, R, Raimi, O.G, Min, J, Zeng, H, Vallius, L, Shepherd, S, Ibrahim, A.F.M, Wu, H, Plotnikov, A.N, van Aalten, D.M.F. | | Deposit date: | 2008-07-05 | | Release date: | 2008-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Kinetic Differences between Human and Aspergillus Fumigatus D-Glucosamine-6- Phosphate N-Acetyltransferase.
Biochem.J., 415, 2008
|
|
2VUR
 
 | | Chemical dissection of the link between Streptozotocin, O-GlcNAc and pancreatic cell death | | Descriptor: | 2-deoxy-2-{[(2-hydroxy-1-methylhydrazino)carbonyl]amino}-beta-D-glucopyranose, O-GLCNACASE NAGJ, SULFATE ION | | Authors: | Pathak, S, Dorfmueller, H.C, Borodkin, V.S, van Aalten, D.M.F. | | Deposit date: | 2008-05-29 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical Dissection of the Link between Streptozotocin, O-Glcnac, and Pancreatic Cell Death.
Chem.Biol., 15, 2008
|
|
2WB5
 
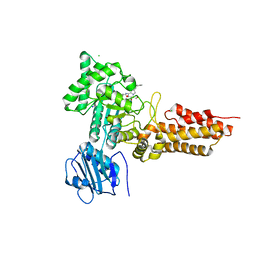 | | GlcNAcstatins are nanomolar inhibitors of human O-GlcNAcase inducing cellular hyper-O-GlcNAcylation | | Descriptor: | (5R,6R,7R,8S)-6,7-dihydroxy-5-(hydroxymethyl)-2-(2-phenylethyl)-8-(propanoylamino)-5,6,7,8-tetrahydro-1H-imidazo[1,2-a]pyridin-4-ium, CHLORIDE ION, O-GLCNACASE NAGJ, ... | | Authors: | Dorfmueller, H.C, Borodkin, V.S, Schimpl, M, van Aalten, D.M.F. | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Glcnacstatins are Nanomolar Inhibitors of Human O-Glcnacase Inducing Cellular Hyper-O-Glcnacylation
Biochem.J., 420, 2009
|
|
2VYO
 
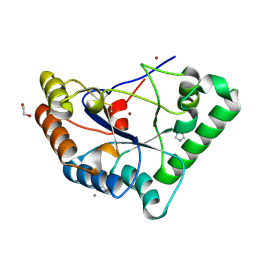 | | Chitin deacetylase family member from Encephalitozoon cuniculi | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Urch, J.E, Hurtado-Guerrero, R, Texier, C, Van Aalten, D.M.F. | | Deposit date: | 2008-07-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of a Putative Polysaccharide Deacetylase of the Human Parasite Encephalitozoon Cuniculi.
Protein Sci., 18, 2009
|
|
2UVM
 
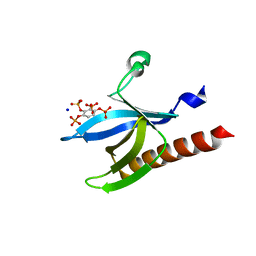 | | Structure of PKBalpha PH domain in complex with a novel inositol headgroup surrogate, benzene 1,2,3,4-tetrakisphosphate | | Descriptor: | BENZENE-1,2,3,4-TETRAYL TETRAKIS[DIHYDROGEN (PHOSPHATE)], RAC-ALPHA SERINE/THREONINE-PROTEIN KINASE, SODIUM ION | | Authors: | Komander, D, Mills, S.J, Trusselle, M.N, Safrany, S.T, van Aalten, D.M.F, Potter, B.V.L. | | Deposit date: | 2007-03-12 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel Inositol Phospholipid Headgroup Surrogate Crystallised in the Pleckstrin Homology Domain of Protein Kinase Balpha.
Acs Chem.Biol., 2, 2007
|
|
2UY3
 
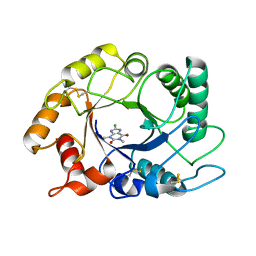 | |
2W63
 
 | |
2W61
 
 | |
2W62
 
 | | Saccharomyces cerevisiae Gas2p in complex with laminaripentaose | | Descriptor: | 1,4-BUTANEDIOL, GLYCOLIPID-ANCHORED SURFACE PROTEIN 2, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Schuettelkopf, A.W, Hurtado-Guerrero, R, van Aalten, D.M.F. | | Deposit date: | 2008-12-16 | | Release date: | 2009-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Mechanisms of Yeast Cell Wall Glucan Remodeling.
J.Biol.Chem., 284, 2009
|
|
