6CET
 
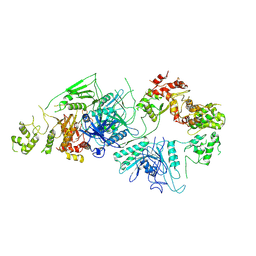 | | Cryo-EM structure of GATOR1 | | Descriptor: | GATOR complex protein DEPDC5, GATOR complex protein NPRL2, GATOR complex protein NPRL3 | | Authors: | Shen, K, Huang, R.K, Brignole, E.J, Yu, Z, Sabatini, D.M. | | Deposit date: | 2018-02-12 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes.
Nature, 556, 2018
|
|
2VVY
 
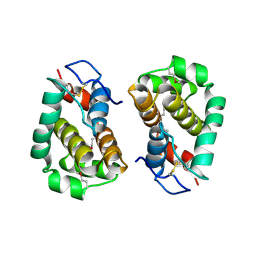 | | Structure of Vaccinia virus protein B14 | | Descriptor: | PROTEIN B15 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
3FB6
 
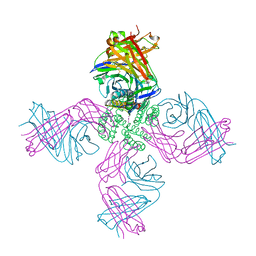 | | KcsA Potassium channel in the partially open state with 16 A opening at T112 | | Descriptor: | POTASSIUM ION, Voltage-gated potassium channel, antibody fab fragment heavy chain, ... | | Authors: | Cuello, L.G, Jogini, V, Cortes, D.M, Perozo, E. | | Deposit date: | 2008-11-18 | | Release date: | 2010-05-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | KcsA Potassium channel in the partially open state with 16 A opening at T112
TO BE PUBLISHED
|
|
1E3Z
 
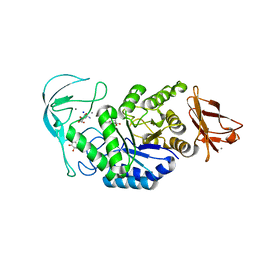 | | Acarbose complex of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.93A | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
6BY3
 
 | | Open and conductive conformation of KcsA-T75A mutant | | Descriptor: | Antibody Heavy Chain, Antibody Light Chain, DIACYL GLYCEROL, ... | | Authors: | Labro, A.J, Cortes, D.M, Tilegenova, C, Cuello, L.G. | | Deposit date: | 2017-12-19 | | Release date: | 2018-05-09 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Inverted allosteric coupling between activation and inactivation gates in K+channels.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C6A
 
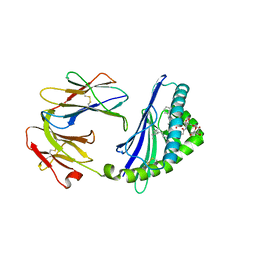 | | Structure of glycolipid aGSA[16,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6C6F
 
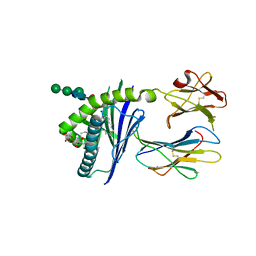 | |
2UYJ
 
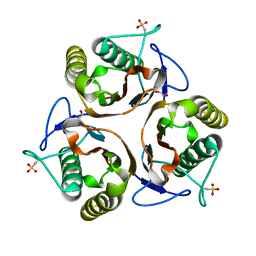 | | Crystal structure of E. coli TdcF with bound ethylene glycol | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN TDCF | | Authors: | Burman, J.D, Stevenson, C.E.M, Sawers, R.G, Lawson, D.M. | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Crystal Structure of Escherichia Coli Tdcf, a Member of the Highly Conserved Yjgf/Yer057C/Uk114 Family.
Bmc Struct.Biol., 7, 2007
|
|
2UYP
 
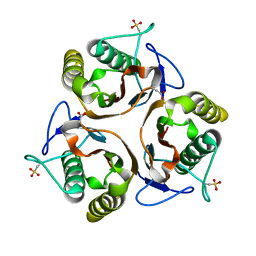 | | Crystal structure of E. coli TdcF with bound propionate | | Descriptor: | PROPANOIC ACID, PROTEIN TDCF | | Authors: | Burman, J.D, Stevenson, C.E.M, Sawers, R.G, Lawson, D.M. | | Deposit date: | 2007-04-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The Crystal Structure of Escherichia Coli Tdcf, a Member of the Highly Conserved Yjgf/Yer057C/Uk114 Family.
Bmc Struct.Biol., 7, 2007
|
|
1F6G
 
 | | POTASSIUM CHANNEL (KCSA) FULL-LENGTH FOLD | | Descriptor: | VOLTAGE-GATED POTASSIUM CHANNEL | | Authors: | Cortes, D.M, Perozo, E. | | Deposit date: | 2000-06-21 | | Release date: | 2001-02-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating.
J.Gen.Physiol., 117, 2001
|
|
2UYB
 
 | | S161A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, MANGANESE (II) ION, ... | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
3FN1
 
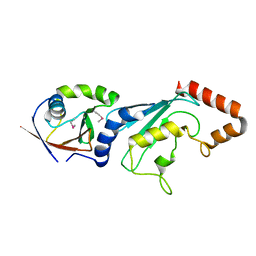 | | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. | | Descriptor: | NEDD8-activating enzyme E1 catalytic subunit, NEDD8-conjugating enzyme UBE2F | | Authors: | Huang, D.T, Ayrault, O, Hunt, H.W, Taherbhoy, A.M, Duda, D.M, Scott, D.C, Borg, L.A, Neale, G, Murray, P.J, Roussel, M.F, Schulman, B.A. | | Deposit date: | 2008-12-22 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification
Mol.Cell, 33, 2009
|
|
2UYA
 
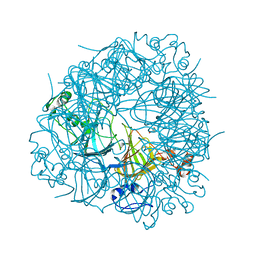 | | DEL162-163 mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
1FD4
 
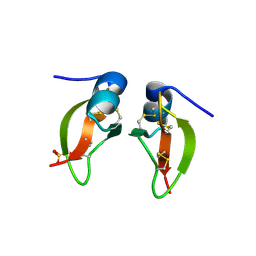 | | HUMAN BETA-DEFENSIN 2 | | Descriptor: | BETA-DEFENSIN 2, SULFATE ION | | Authors: | Hoover, D.M, Lubkowski, J. | | Deposit date: | 2000-07-19 | | Release date: | 2000-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of human beta-defensin-2 shows evidence of higher order oligomerization.
J.Biol.Chem., 275, 2000
|
|
3F0Y
 
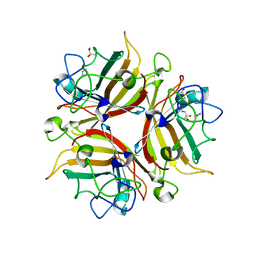 | | Crystal structure of the human Adenovirus type 14 fiber knob | | Descriptor: | Fiber protein, GLYCEROL, IMIDAZOLE | | Authors: | Persson, B.D, Reiter, D.M, Arnberg, N, Stehle, T. | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An arginine switch in the species B adenovirus knob determines high-affinity engagement of cellular receptor CD46
J.Virol., 83, 2009
|
|
1FBL
 
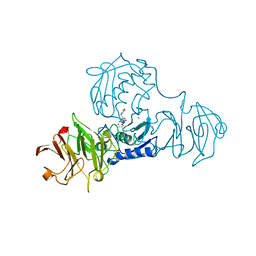 | | STRUCTURE OF FULL-LENGTH PORCINE SYNOVIAL COLLAGENASE (MMP1) REVEALS A C-TERMINAL DOMAIN CONTAINING A CALCIUM-LINKED, FOUR-BLADED BETA-PROPELLER | | Descriptor: | CALCIUM ION, FIBROBLAST (INTERSTITIAL) COLLAGENASE (MMP-1), N-[3-(N'-HYDROXYCARBOXAMIDO)-2-(2-METHYLPROPYL)-PROPANOYL]-O-TYROSINE-N-METHYLAMIDE, ... | | Authors: | Li, J, Brick, P, Blow, D.M. | | Deposit date: | 1995-04-24 | | Release date: | 1996-01-29 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of full-length porcine synovial collagenase reveals a C-terminal domain containing a calcium-linked, four-bladed beta-propeller.
Structure, 3, 1995
|
|
1FD3
 
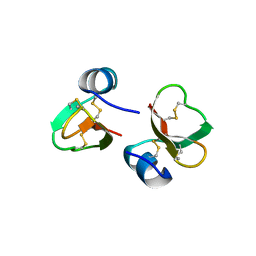 | | HUMAN BETA-DEFENSIN 2 | | Descriptor: | BETA-DEFENSIN 2, SULFATE ION | | Authors: | Hoover, D.M, Lubkowski, J. | | Deposit date: | 2000-07-19 | | Release date: | 2000-11-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of human beta-defensin-2 shows evidence of higher order oligomerization.
J.Biol.Chem., 275, 2000
|
|
6C69
 
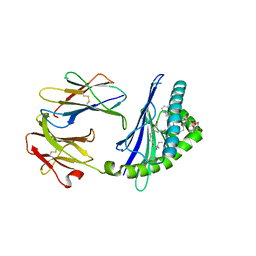 | | Structure of glycolipid aGSA[12,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6C6E
 
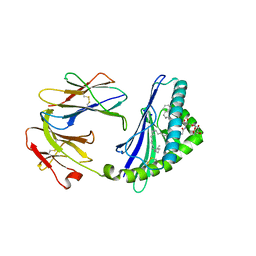 | | Structure of glycolipid aGSA[26,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
2W1E
 
 | | Structure determination of Aurora Kinase in complex with inhibitor | | Descriptor: | 4-[(2-{4-[(PHENYLCARBAMOYL)AMINO]-1H-PYRAZOL-3-YL}-1H-BENZIMIDAZOL-5-YL)METHYL]MORPHOLIN-4-IUM, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Howard, S, Berdini, V, Boulstridge, J.A, Carr, M.G, Cross, D.M, Curry, J, Devine, L.A, Early, T.R, Fazal, L, Gill, A.L, Heathcote, M, Maman, S, Matthews, J.E, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Rees, D.C, Reule, M, Tisi, D, Williams, G, Vinkovic, M, Wyatt, P.G. | | Deposit date: | 2008-10-17 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Fragment-Based Discovery of the Pyrazol-4-Yl Urea (at9283), a Multitargeted Kinase Inhibitor with Potent Aurora Kinase Activity.
J.Med.Chem., 52, 2009
|
|
2VTM
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, PYRAZOLO[1,5-A]PYRIMIDINE-3-CARBONITRILE | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
6D3N
 
 | | Crystal structure of h4-1BB ligand | | Descriptor: | GLYCEROL, Tumor necrosis factor ligand superfamily member 9 | | Authors: | Aruna, B, Zajonc, D.M, Doukov, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the human 4-1BB receptor bound to its ligand 4-1BBL reveal covalent receptor dimerization as a potential signaling amplifier.
J. Biol. Chem., 293, 2018
|
|
2VU3
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | 4-{[(2,6-dichlorophenyl)carbonyl]amino}-N-piperidin-4-yl-1H-pyrazole-3-carboxamide, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-20 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VV9
 
 | | CDK2 in complex with an imidazole piperazine | | Descriptor: | 2-{4-[4-({4-[2-methyl-1-(1-methylethyl)-1H-imidazol-5-yl]pyrimidin-2-yl}amino)phenyl]piperazin-1-yl}-2-oxoethanol, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Acton, D.G, Anderson, M, Andrews, D.M, Barker, A.J, Brassington, C.A, Finlay, M.R, Fisher, E, Gerhardt, S, Graham, M.A, Green, C.P, Heaton, D.W, Loddick, S.A, Morgentin, R, Read, J, Roberts, A, Stanway, J, Tucker, J.A, Weir, H.M. | | Deposit date: | 2008-06-04 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Imidazole Piperazines: Sar and Development of a Potent Class of Cyclin-Dependent Kinase Inhibitors with a Novel Binding Mode.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VVX
 
 | | Structure of Vaccinia virus protein A52 | | Descriptor: | PROTEIN A52 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
