6MIV
 
 | | Crystal structure of the mCD1d/xxq (JJ300)/iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
6MJA
 
 | | Crystal structure of the mCD1d/xxo (JJ294) /iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
4IBK
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(2S)-2-(7-chloro-1,3-benzodioxol-5-yl)-3-[(5-chlorothiophen-2-yl)carbonyl]-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
2VTJ
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design | | Descriptor: | 4-[(6-chloropyrazin-2-yl)amino]benzenesulfonamide, CELL DIVISION PROTEIN KINASE 2, GLYCEROL | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VTI
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-(4-sulfamoylphenyl)-1H-indazole-3-carboxamide | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VTR
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design | | Descriptor: | 5-chloro-7-[(1-methylethyl)amino]pyrazolo[1,5-a]pyrimidine-3-carbonitrile, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
6MJQ
 
 | | Crystal structure of the mCD1d/xxp (JJ295) /iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-21 | | Release date: | 2019-01-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
4IBC
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | DIMETHYL SULFOXIDE, Polymerase cofactor VP35, {4-[(2R)-3-(2-chlorobenzoyl)-2-(2-chlorophenyl)-4-hydroxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl]phenyl}acetic acid | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.745 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
1E43
 
 | | Native structure of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.7A | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
6MJI
 
 | | Crystal structure of the mCD1d/xxs (JJ304) /iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
6MKB
 
 | | Crystal structure of murine 4-1BB ligand | | Descriptor: | SODIUM ION, SULFATE ION, Tumor necrosis factor ligand superfamily member 9, ... | | Authors: | Bitra, A, Zajonc, D.M, Doukov, T. | | Deposit date: | 2018-09-25 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the m4-1BB/4-1BBL complex reveals an unusual dimeric ligand that undergoes structural changes upon 4-1BB receptor binding.
J. Biol. Chem., 294, 2019
|
|
6MKZ
 
 | | Crystal structure of murine 4-1BB/4-1BBL complex | | Descriptor: | Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor receptor superfamily member 9, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bitra, A, Zajonc, D.M, Doukov, T. | | Deposit date: | 2018-09-26 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the m4-1BB/4-1BBL complex reveals an unusual dimeric ligand that undergoes structural changes upon 4-1BB receptor binding.
J. Biol. Chem., 294, 2019
|
|
8V2F
 
 | | Crystal structure of IRAK4 kinase domain with compound 9 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
7W9A
 
 | |
7W6S
 
 | | CryoEM structure of human KChIP2-Kv4.3 complex | | Descriptor: | Isoform 2 of Potassium voltage-gated channel subfamily D member 3, Kv channel-interacting protein 2 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
8V2L
 
 | | Crystal structure of IRAK4 kinase domain with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, Interleukin-1 receptor-associated kinase 4, N-{2-[4-(hydroxymethyl)phenyl]-6-(2-hydroxypropan-2-yl)-2H-indazol-5-yl}-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
7W6N
 
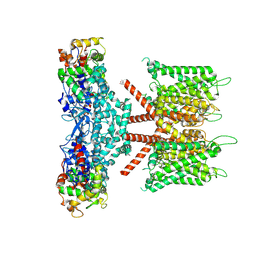 | | CryoEM structure of human KChIP1-Kv4.3 complex | | Descriptor: | Isoform 2 of Potassium voltage-gated channel subfamily D member 3, Kv channel-interacting protein 1 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
7W6T
 
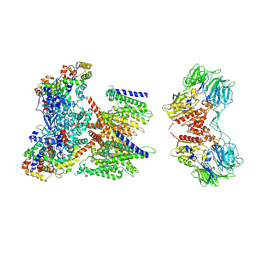 | | CryoEM structure of human KChIP1-Kv4.3-DPP6 complex | | Descriptor: | Dipeptidyl aminopeptidase-like protein 6, Isoform 2 of Potassium voltage-gated channel subfamily D member 3, Kv channel-interacting protein 1 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
9CCV
 
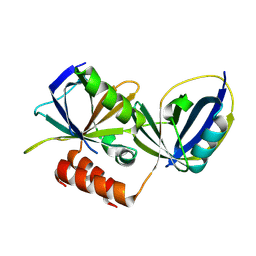 | | Crystal structure of human respiratory syncytial virus NS1 bound to human MED25 ACID | | Descriptor: | Mediator of RNA polymerase II transcription subunit 25, Non-structural protein 1 | | Authors: | Kalita, P, Borek, D.M, Amarasinghe, G.K, Leung, D.W. | | Deposit date: | 2024-06-23 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Molecular basis for human respiratory syncytial virus transcriptional regulator NS1 interactions with MED25.
Nat Commun, 16, 2025
|
|
2XGB
 
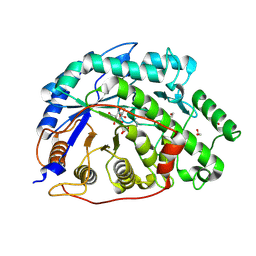 | | Crystal structure of Barley Beta-Amylase complexed with 2,3- epoxypropyl-alpha-D-glucopyranoside | | Descriptor: | (2R)-oxiran-2-ylmethyl alpha-D-glucopyranoside, 1,2-ETHANEDIOL, BETA-AMYLASE | | Authors: | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | Deposit date: | 2010-06-02 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Chemical Genetics and Cereal Starch Metabolism: Structural Basis of the Non-Covalent and Covalent Inhibition of Barley Beta-Amylase.
Mol.Biosyst., 7, 2011
|
|
8V1O
 
 | | Crystal structure of IRAK4 kinase domain with compound 4 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
2XGI
 
 | | Crystal structure of Barley Beta-Amylase complexed with 3,4- epoxybutyl alpha-D-glucopyranoside | | Descriptor: | (3R)-3-hydroxybutyl alpha-D-glucopyranoside, (3S)-3-hydroxybutyl alpha-D-glucopyranoside, 1,2-ETHANEDIOL, ... | | Authors: | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | Deposit date: | 2010-06-04 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Chemical genetics and cereal starch metabolism: structural basis of the non-covalent and covalent inhibition of barley beta-amylase.
Mol Biosyst, 7, 2011
|
|
8Y8M
 
 | | Crystal structure of a benzaldehyde lyase mutant M3 from Herbiconiux sp. SALV-R1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Li, Y, Zhang, Y.F, Chen, Y.Y, Liu, W.D, Yao, P.Y, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2024-02-06 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Manipulating Activity and Chemoselectivity of a Benzaldehyde Lyase for Efficient Synthesis of alpha-Hydroxymethyl Ketones and One-Pot Enantio-Complementary Conversion to 1,2-Diols
Acs Catalysis, 14, 2024
|
|
6MIY
 
 | | Crystal structure of the mCD1d/xxa (JJ239)/iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
6MJJ
 
 | | Crystal structure of the mCD1d/xxm (JJ290) /iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-21 | | Release date: | 2019-01-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
