1PCW
 
 | | Aquifex aeolicus KDO8PS in complex with cadmium and APP, a bisubstrate inhibitor | | Descriptor: | 1-DEOXY-6-O-PHOSPHONO-1-[(PHOSPHONOMETHYL)AMINO]-L-THREO-HEXITOL, 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION | | Authors: | Xu, X, Wang, J, Grison, C, Petek, S, Coutrot, P, Birck, M, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2003-05-17 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design of Novel Inhibitors of 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase.
Drug DES.DISCOVERY, 18, 2003
|
|
1PH3
 
 | |
1PH4
 
 | |
1PH5
 
 | |
1PH7
 
 | |
1OJJ
 
 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7BE197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
1P4O
 
 | | Structure of Apo unactivated IGF-1R KInase domain at 1.5A resolution. | | Descriptor: | Insulin-like growth factor I receptor protein | | Authors: | Munshi, S, Kornienko, M, Hall, D.L, Darke, P.L, Waxman, L, Kuo, L.C. | | Deposit date: | 2003-04-23 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of apo, unactivated insulin-like growth factor-1 receptor kinase at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1Q5A
 
 | | S-shaped trans interactions of cadherins model based on fitting C-cadherin (1L3W) to 3D map of desmosomes obtained by electron tomography | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, W, Cowin, P, Stokes, D.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Untangling Desmosomal Knots with Electron Tomography
Science, 302, 2003
|
|
1PA6
 
 | |
1Q55
 
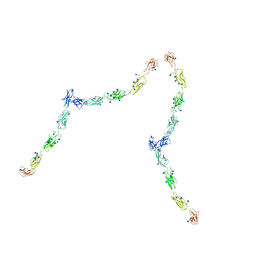 | | W-shaped trans interactions of cadherins model based on fitting C-cadherin (1L3W) to 3D map of desmosomes obtained by electron tomography | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, W, Cowin, P, Stokes, D.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Untangling Desmosomal Knots with Electron Tomography
Science, 302, 2003
|
|
1Q5C
 
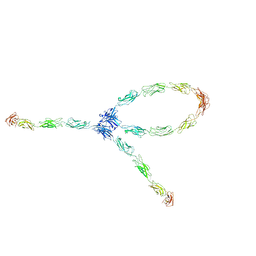 | | S-S-lambda-shaped TRANS and CIS interactions of cadherins model based on fitting C-cadherin (1L3W) to 3D map of desmosomes obtained by electron tomography | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, W, Cowin, P, Stokes, D.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Untangling Desmosomal Knots with Electron Tomography
Science, 302, 2003
|
|
1PJL
 
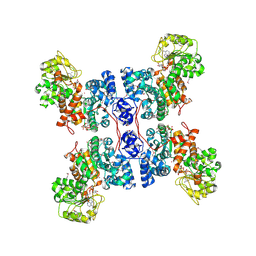 | | Crystal structure of human m-NAD-ME in ternary complex with NAD and Lu3+ | | Descriptor: | LUTETIUM (III) ION, NAD-dependent malic enzyme, mitochondrial, ... | | Authors: | Yang, Z, Batra, R, Floyd, D.L, Hung, H.-C, Chang, G.-G, Tong, L. | | Deposit date: | 2003-06-03 | | Release date: | 2003-06-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Potent and competitive inhibition of malic enzymes by lanthanide ions
Biochem.Biophys.Res.Commun., 274, 2000
|
|
1Q1M
 
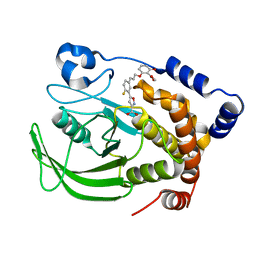 | | A Highly Efficient Approach to a Selective and Cell Active PTP1B inhibitors | | Descriptor: | 5-{2-FLUORO-5-[3-(3-HYDROXY-2-METHOXYCARBONYL-PHENOXY)-PROPENYL]-PHENYL}-ISOXAZOLE-3-CARBOXYLIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Liu, G, Xin, Z, Pei, Z, Hajduk, P.J, Abad-Zapatero, C, Hutchins, C.W, Zhao, H, Lubben, T.H, Ballaron, S.J, Haasch, D.L, Kaszubska, W, Rondinone, C.M, Trevillyan, J.M, Jirousek, M.R. | | Deposit date: | 2003-07-22 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment screening and assembly: a highly efficient approach to a selective and cell active protein tyrosine phosphatase 1B inhibitor.
J.Med.Chem., 46, 2003
|
|
1POD
 
 | |
1Q5B
 
 | | lambda-shaped TRANS and CIS interactions of cadherins model based on fitting C-cadherin (1L3W) to 3D map of desmosomes obtained by electron tomography | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, W, Cowin, P, Stokes, D.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Untangling Desmosomal Knots with Electron Tomography
Science, 302, 2003
|
|
4R5J
 
 | | Crystal structure of the DnaK C-terminus (Dnak-SBD-A) | | Descriptor: | CALCIUM ION, Chaperone protein DnaK, PHOSPHATE ION | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
4R5L
 
 | | Crystal structure of the DnaK C-terminus (Dnak-SBD-C) | | Descriptor: | CALCIUM ION, Chaperone protein DnaK, PHOSPHATE ION, ... | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9701 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
4R5G
 
 | | Crystal structure of the DnaK C-terminus with the inhibitor PET-16 | | Descriptor: | Chaperone protein DnaK, triphenyl(phenylethynyl)phosphonium | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4501 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
4R5I
 
 | | Crystal structure of the DnaK C-terminus with the substrate peptide NRLLLTG | | Descriptor: | Chaperone protein DnaK, HSP70/DnaK Substrate Peptide: NRLLLTG, PHOSPHATE ION, ... | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9702 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
1RER
 
 | | Crystal structure of the homotrimer of fusion glycoprotein E1 from Semliki Forest Virus. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gibbons, D.L, Vaney, M.C, Roussel, A, Vigouroux, A, Reilly, B, Kielian, M, Rey, F.A. | | Deposit date: | 2003-11-07 | | Release date: | 2004-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus.
Nature, 427, 2004
|
|
4RN5
 
 | | B1 domain of human Neuropilin-1 with acetate ion in a ligand-binding site | | Descriptor: | ACETATE ION, GLYCEROL, Neuropilin-1, ... | | Authors: | Allerston, C.K, Yelland, T.S, Jarvis, A, Jenkins, K, Winfield, N, Cheng, L, Jia, H, Zachary, I, Selwood, D.L, Djordjevic, S. | | Deposit date: | 2014-10-23 | | Release date: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Conserved water molecules in a ligand-binding site of neuropilin-1
To be Published
|
|
4U2C
 
 | | Crystal structure of dienelactone hydrolase A-6 variant (S7T, A24V, Q35H, F38L, Q110L, C123S, Y145C, E199G and S208G) at 1.95 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4U2D
 
 | | Crystal structure of dienelactone hydrolase S-2 variant (Q35H, F38L, Q110L, C123S, Y137C, Y145C, N154D, E199G, S208G and G211D) at 1.67 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4U2G
 
 | | Crystal structure of dienelactone hydrolase B-4 variant (Q35H, F38L, Y64H, Q76L, Q110L, C123S, Y137C, A141V, Y145C, N154D, E199G, S208G, G211D, S233G and 237Q) at 1.80 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
7TYL
 
 | | Calcitonin Receptor in complex with Gs and rat amylin peptide, bypass motif | | Descriptor: | Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-13 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
