2O93
 
 | |
2NUE
 
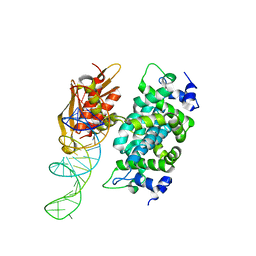 | | Crystal structure of RNase III from Aquifex aeolicus complexed with ds-RNA at 2.9-Angstrom Resolution | | Descriptor: | 46-MER, Ribonuclease III | | Authors: | Gan, J.H, Shaw, G, Tropea, J.E, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2006-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A stepwise model for double-stranded RNA processing by ribonuclease III.
Mol.Microbiol., 67, 2007
|
|
2NXG
 
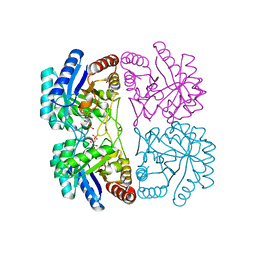 | | Structural and mechanistic changes along an engineered path from metallo to non-metallo KDO8P synthase. | | Descriptor: | (2R,4R,5R,6R,7R)-2,4,5,6,7-PENTAHYDROXY-2,8-BIS(PHOSPHONOOXY)OCTANOIC ACID, 2-dehydro-3-deoxyphosphooctonate aldolase, ARABINOSE-5-PHOSPHATE, ... | | Authors: | Kona, F, Xu, X, Martin, P, Kuzmic, P, Gatti, D.L. | | Deposit date: | 2006-11-17 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Mechanistic Changes along an Engineered Path from Metallo to Nonmetallo 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthases.
Biochemistry, 46, 2007
|
|
2NX3
 
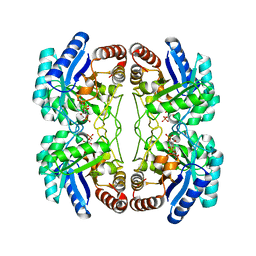 | | Structural and mechanistic changes along an engineered path from metallo to non-metallo KDO8P synthase | | Descriptor: | (2R,4R,5R,6R,7R)-2,4,5,6,7-PENTAHYDROXY-2,8-BIS(PHOSPHONOOXY)OCTANOIC ACID, 2-dehydro-3-deoxyphosphooctonate aldolase, ARABINOSE-5-PHOSPHATE, ... | | Authors: | Kona, F, Xu, X, Martin, P, Kuzmic, P, Gatti, D.L. | | Deposit date: | 2006-11-16 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Changes along an Engineered Path from Metallo to Nonmetallo 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthases.
Biochemistry, 46, 2007
|
|
2OVC
 
 | |
2P4F
 
 | |
2P4X
 
 | |
2L4T
 
 | | GIP/Glutaminase L peptide complex | | Descriptor: | Glutaminase L peptide, Tax1-binding protein 3 | | Authors: | Zoetewey, D.L, Ovee, M, Banerjee, M, Bhaskaran, R, Mohanty, S. | | Deposit date: | 2010-10-13 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Promiscuous binding at the crossroads of numerous cancer pathways: insight from the binding of glutaminase interacting protein with glutaminase L.
Biochemistry, 50, 2011
|
|
2L4K
 
 | | Water refined solution structure of the human Grb7-SH2 domain in complex with the 10 amino acid peptide pY1139 | | Descriptor: | Growth factor receptor-bound protein 7, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Pias, S.C, Ivancic, M, Brescia, P.J, Johnson, D.L, Smith, D.E, Daly, R.J, Lyons, B.A. | | Deposit date: | 2010-10-07 | | Release date: | 2010-11-17 | | Last modified: | 2012-10-03 | | Method: | SOLUTION NMR | | Cite: | Water-Refined Solution Structure of the Human Grb7-SH2 Domain in Complex with the erbB2 Receptor Peptide pY1139.
Protein Pept.Lett., 19, 2012
|
|
2L4S
 
 | | Promiscuous Binding at the Crossroads of Numerous Cancer Pathways: Insight from the Binding of GIP with Glutaminase L | | Descriptor: | Tax1-binding protein 3 | | Authors: | Zoetewey, D.L, Ovee, M, Banerjee, M, Bhaskaran, R, Mohanty, S. | | Deposit date: | 2010-10-13 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous binding at the crossroads of numerous cancer pathways: insight from the binding of glutaminase interacting protein with glutaminase L.
Biochemistry, 50, 2011
|
|
2MS3
 
 | |
2LZZ
 
 | | Solution structure of a mutant of the triheme cytochrome PpcA from Geobacter sulfurreducens sheds light on the role of the conserved aromatic residue F15 | | Descriptor: | Cytochrome c, 3 heme-binding sites, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dantas, J.M, Morgado, L, Turner, D.L, Salgueiro, C.A. | | Deposit date: | 2012-10-12 | | Release date: | 2013-01-30 | | Last modified: | 2013-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a mutant of the triheme cytochrome PpcA from Geobacter sulfurreducens sheds light on the role of the conserved aromatic residue F15.
Biochim.Biophys.Acta, 1827, 2013
|
|
2Q99
 
 | |
7Y78
 
 | | Crystal structure of Cry78Aa | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Toxin | | Authors: | Cao, B.B, Nie, Y.F, Wang, N.C, Guan, Z.Y, Zhang, D.L, Zhang, J. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of Cry78Aa from Bacillus thuringiensis provides insights into its insecticidal activity.
Commun Biol, 5, 2022
|
|
7Y79
 
 | | Crystal structure of Cry78Aa | | Descriptor: | Toxin | | Authors: | Cao, B.B, Nie, Y.F, Wang, N.C, Guan, Z.Y, Zhang, D.L, Zhang, J. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The crystal structure of Cry78Aa from Bacillus thuringiensis provides insights into its insecticidal activity.
Commun Biol, 5, 2022
|
|
7M2M
 
 | | NMR Structure of GCAP5 | | Descriptor: | Guanylate cyclase activator 1A, MAGNESIUM ION | | Authors: | Ames, J.B, Cudia, D.L. | | Deposit date: | 2021-03-17 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | NMR and EPR-DEER Structure of a Dimeric Guanylate Cyclase Activator Protein-5 from Zebrafish Photoreceptors.
Biochemistry, 60, 2021
|
|
7Y99
 
 | | Crystal Structure Analysis of cp2 bound BCLxl | | Descriptor: | Bcl-2-like protein 1, CP2 peptide, N-(2-acetamidoethyl)-4-(4,5-dihydro-1,3-thiazol-2-yl)benzamide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
1YKJ
 
 | | A45G p-hydroxybenzoate hydroxylase with p-hydroxybenzoate bound | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, P-hydroxybenzoate hydroxylase, ... | | Authors: | Cole, L.J, Gatti, D.L, Entsch, B, Ballou, D.P. | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Removal of a methyl group causes global changes in p-hydroxybenzoate hydroxylase.
Biochemistry, 44, 2005
|
|
1VZE
 
 | | L. CASEI THYMIDYLATE SYNTHASE MUTANT E60Q TERNARY COMPLEX WITH DUMP AND CB3717 | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Birdsall, D.L, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1996-09-18 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The separate effects of E60Q in Lactobacillus casei thymidylate synthase delineate between mechanisms for formation of intermediates in catalysis.
Protein Eng., 11, 1998
|
|
1VZD
 
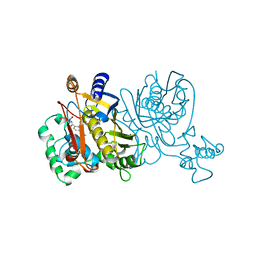 | | L. CASEI THYMIDYLATE SYNTHASE MUTANT E60Q TERNARY COMPLEX WITH FDUMP AND CB3717 | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Birdsall, D.L, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1996-09-18 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The separate effects of E60Q in Lactobacillus casei thymidylate synthase delineate between mechanisms for formation of intermediates in catalysis.
Protein Eng., 11, 1998
|
|
1VZA
 
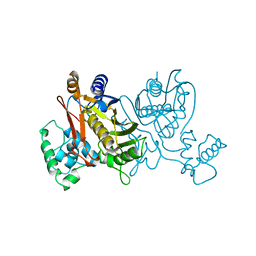 | |
1VZC
 
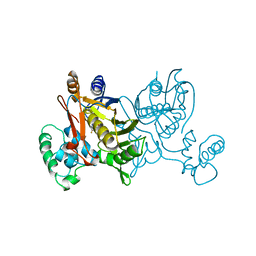 | |
7YAA
 
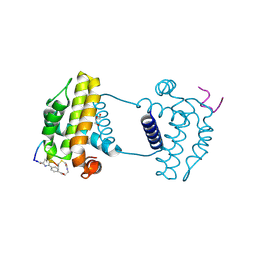 | | Crystal structure analysis of cp3 bound BCLxl | | Descriptor: | Bcl-2-like protein 1, GLYCEROL, N-(2-acetamidoethyl)-4-(4-methanoyl-1,3-thiazol-2-yl)benzamide, ... | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7Y8D
 
 | | Crystal structure of cp1 bound BCLxl | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Bcl-2-like protein 1, cp1 peptide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7YA5
 
 | | Crystal structure analysis of cp1 bound BCL2/G101V | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Apoptosis regulator Bcl-2, cp1 peptide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
