7M1A
 
 | | SusE-like protein BT2857 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Galactose-binding like protein | | Authors: | Suits, M.D.L. | | Deposit date: | 2021-03-12 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Analysis of Two SusE-Like Enzymes From Bacteroides thetaiotaomicron Reveals a Potential Degradative Capacity for This Protein Family.
Front Microbiol, 12, 2021
|
|
5WUC
 
 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | SODIUM ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
7M1B
 
 | | SusE-like protein BT2857 | | Descriptor: | 1,2-ETHANEDIOL, Galactose-binding-like protein | | Authors: | Suits, M.D.L. | | Deposit date: | 2021-03-12 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Analysis of Two SusE-Like Enzymes From Bacteroides thetaiotaomicron Reveals a Potential Degradative Capacity for This Protein Family.
Front Microbiol, 12, 2021
|
|
2Q99
 
 | |
5WUD
 
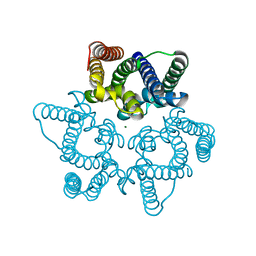 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
7MQP
 
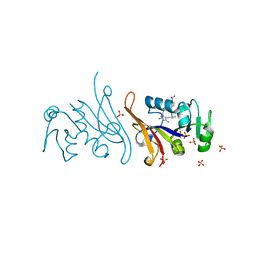 | | E. coli dihydrofolate reductase complexed with 4'-chloro-3'-(4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl)-[1,1'-biphenyl]-4-carboxamide (UCP1228) | | Descriptor: | 3'-[(2S)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-4'-methyl[1,1'-biphenyl]-4-carboxamide, Dihydrofolate reductase, SULFATE ION | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
2QFG
 
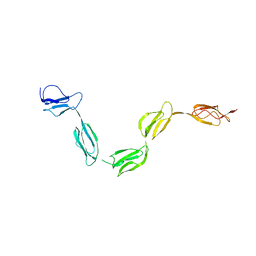 | | Solution Structure of the N-terminal SCR-1/5 fragment of Complement Factor H. | | Descriptor: | Complement factor H | | Authors: | Okemefuna, A.I, Gilbert, H.E, Griggs, K.M, Ormsby, R.J, Gordon, D.L, Perkins, S.J. | | Deposit date: | 2007-06-27 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | The regulatory SCR-1/5 and cell surface-binding SCR-16/20 fragments of factor H reveal partially folded-back solution structures and different self-associative properties.
J.Mol.Biol., 375, 2008
|
|
7MJS
 
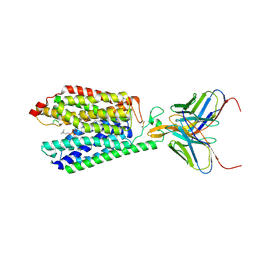 | | Single-Particle Cryo-EM Structure of Major Facilitator Superfamily Domain containing 2A in complex with LPC-18:3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2AG3 Fab heavy chain, 2AG3 Fab light chain, ... | | Authors: | Cater, R.J, Chua, G.L, Erramilli, S.K, Keener, J.E, Choy, B.C, Tokarz, P, Chin, C.F, Quek, D.Q.Y, Kloss, B, Pepe, J.G, Parisi, G, Wong, B.H, Clarke, O.B, Marty, M.T, Kossiakoff, A.A, Khelashvili, G, Silver, D.L, Mancia, F. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis of omega-3 fatty acid transport across the blood-brain barrier.
Nature, 595, 2021
|
|
5WUE
 
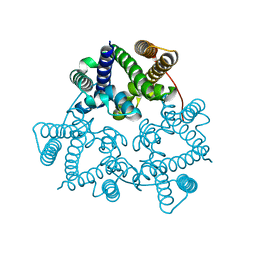 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
2R1A
 
 | |
4XYJ
 
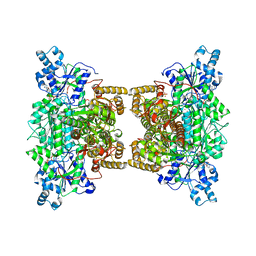 | | Crystal structure of human phosphofructokinase-1 in complex with ATP and Mg, Northeast Structural Genomics Consortium Target HR9275 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent 6-phosphofructokinase, platelet type, ... | | Authors: | Forouhar, F, Webb, B.A, Szu, F.-E, Seetharaman, J, Barber, D.L, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-02-02 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations.
Nature, 523, 2015
|
|
7NPX
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 3-(3-Methoxyquinoxalin-2-yl)propanoic acid at 24 hours of soaking | | Descriptor: | 3-(3-methoxyquinoxalin-2-yl)propanoic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fata, F, Silvestri, I, Williams, D.L, Angelucci, F. | | Deposit date: | 2021-02-28 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Probing the Surface of a Parasite Drug Target Thioredoxin Glutathione Reductase Using Small Molecule Fragments.
Acs Infect Dis., 7, 2021
|
|
4XVZ
 
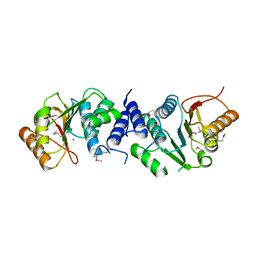 | | MycF mycinamicin III 3'-O-methyltransferase in complex with Mg | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mycinamicin III 3''-O-methyltransferase | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2015-01-28 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
1DSI
 
 | | Solution structure of a duocarmycin sa-indole-alkylated dna dupleX | | Descriptor: | 4-HYDROXY-6-(1H-INDOLE-2-CARBONYL)-8-METHYL-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3') | | Authors: | Schnell, J.R, Ketchem, R.R, Boger, D.L, Chazin, W.J. | | Deposit date: | 1998-07-29 | | Release date: | 1998-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Binding-Induced Activation of DNA Alkylation by Duocarmycin SA: Insights from the Structure of an Indole Derivative-DNA Adduct
J.Am.Chem.Soc., 121, 1999
|
|
6E3Y
 
 | | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor | | Descriptor: | Calcitonin gene-related peptide 1, Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liang, Y.L, Khoshouei, M, Deganutti, G, Glukhova, A, Koole, C, Peat, T.S, Radjainia, M, Plitzko, J.M, Baumeister, W, Miller, L.J, Hay, D.L, Christopoulos, A, Reynolds, C.A, Wootten, D, Sexton, P.M. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor.
Nature, 561, 2018
|
|
3ZRC
 
 | | pVHL54-213-EloB-EloC complex (4R)-4-HYDROXY-1-[(3-METHYLISOXAZOL-5-YL)ACETYL]-N-[4-(1,3-OXAZOL-5-YL)BENZYL]-L-PROLINAMIDE bound | | Descriptor: | (4R)-4-HYDROXY-1-[(3-METHYLISOXAZOL-5-YL)ACETYL]-N-[4-(1,3-OXAZOL-5-YL)BENZYL]-L-PROLINAMIDE, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 2, ... | | Authors: | Van Molle, I, Buckley, D.L, Crews, C.M, Ciulli, A. | | Deposit date: | 2011-06-15 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Targeting the Von Hippel-Lindau E3 Ubiquitin Ligase Using Small Molecules to Disrupt the Vhl/Hif-1Alpha Interaction
J.Am.Chem.Soc., 134, 2012
|
|
3ZTD
 
 | | pVHL54-213-EloB-EloC complex _ methyl 4-(((2S,4R)-4-hydroxy-1-(2-(3- methylisoxazol-5-yl)acetyl)pyrrolidine-2-carboxamido)methyl)benzoate | | Descriptor: | METHYL 4-[({(4R)-4-HYDROXY-1-[(3-METHYLISOXAZOL-5-YL)ACETYL]-L-PROLYL}AMINO)METHYL]BENZOATE, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 2, ... | | Authors: | VanMolle, I, Buckley, D.L, Crews, C.M, Ciulli, A. | | Deposit date: | 2011-07-07 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Dissecting Fragment-Based Lead Discovery at the Von Hippel-Lindau Protein:Hypoxia Inducible Factor 1Alpha Protein-Protein Interface.
Chem.Biol., 19, 2012
|
|
1E8E
 
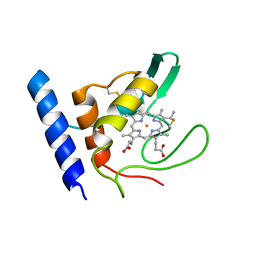 | | Solution Structure of Methylophilus methylotrophus Cytochrome c''. Insights into the Structural Basis of Haem-Ligand Detachment | | Descriptor: | CYTOCHROME C'', HEME C | | Authors: | Brennan, L, Turner, D.L, Fareleira, P, Santos, H. | | Deposit date: | 2000-09-20 | | Release date: | 2001-09-20 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Methylophilus Methylotrophus Cytochrome C": Insights Into the Structural Basis of Haem-Ligand Detachment
J.Mol.Biol., 308, 2001
|
|
8D00
 
 | |
3ZTC
 
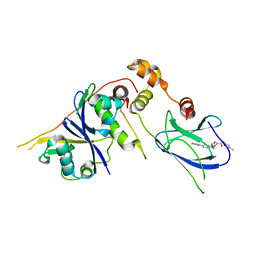 | | pVHL54-213-EloB-EloC complex _ (2S,4R)-N-((1,1'-biphenyl)-4-ylmethyl)- 4-hydroxy-1-(2-(3-methylisoxazol-5-yl)acetyl)pyrrolidine-2- carboxamide | | Descriptor: | (4R)-N-(BIPHENYL-4-YLMETHYL)-4-HYDROXY-1-[(3-METHYLISOXAZOL-5-YL)ACETYL]-L-PROLINAMIDE, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 2, ... | | Authors: | Van Molle, I, Buckley, D.L, Crews, C.M, Ciulli, A. | | Deposit date: | 2011-07-06 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dissecting Fragment-Based Lead Discovery at the Von Hippel-Lindau Protein:Hypoxia Inducible Factor 1Alpha Protein-Protein Interface.
Chem.Biol., 19, 2012
|
|
6EIT
 
 | |
6ESB
 
 | | BK polyomavirus + 20 mM GT1b oligosaccharide | | Descriptor: | CALCIUM ION, Capsid protein VP1, Minor capsid protein VP2, ... | | Authors: | Hurdiss, D.L, Ranson, N.A. | | Deposit date: | 2017-10-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The Structure of an Infectious Human Polyomavirus and Its Interactions with Cellular Receptors.
Structure, 26, 2018
|
|
6EDG
 
 | | Pseudomonas exotoxin A domain III T18H477L | | Descriptor: | Exotoxin, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE | | Authors: | Moss, D.L, Park, H.W, Mettu, R.R, Landry, S.J. | | Deposit date: | 2018-08-09 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Deimmunizing substitutions in Pseudomonasexotoxin domain III perturb antigen processing without eliminating T-cell epitopes.
J.Biol.Chem., 294, 2019
|
|
3ZRF
 
 | | pVHL54-213-EloB-EloC complex_apo | | Descriptor: | TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 2, VON HIPPEL-LINDAU DISEASE TUMOR SUPPRESSOR, | | Authors: | Van Molle, I, Buckley, D.L, Crews, C.M, Ciulli, A. | | Deposit date: | 2011-06-16 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeting the Von Hippel-Lindau E3 Ubiquitin Ligase Using Small Molecules to Disrupt the Vhl/Hif-1Alpha Interaction
J.Am.Chem.Soc., 134, 2012
|
|
8E77
 
 | | rystal structure of Pcryo_0616, the aminotransferase required to synthesize UDP-N-acetyl-3-amino-D-glucosaminuronic acid (UDP-GlcNAc3NA), incomplete with its external aldimine reaction intermediate | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-6-{[(R)-{[(S)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}tetrahydro-2H-pyran-2-carboxylic acid (non-preferred name), 1,2-ETHANEDIOL, DegT/DnrJ/EryC1/StrS aminotransferase, ... | | Authors: | Hofmeister, D.L, Seltzner, C.A, Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-08-23 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
