2YNP
 
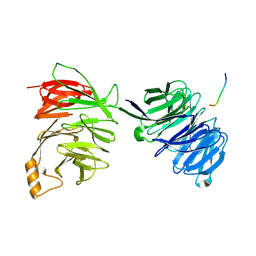 | | yeast betaprime COP 1-604 with KTKTN motif | | Descriptor: | COATOMER SUBUNIT BETA', KTKTN MOTIF | | Authors: | Jackson, L.P, Lewis, M, Kent, H.M, Edeling, M.A, Evans, P.R, Duden, R, Owen, D.J. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.962 Å) | | Cite: | Molecular Basis for Recognition of Dilysine Trafficking Motifs by Copi.
Dev.Cell, 23, 2012
|
|
2Y9Y
 
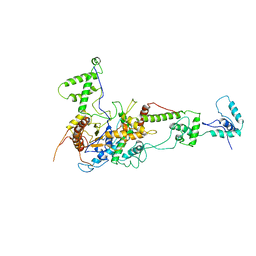 | | Chromatin Remodeling Factor ISW1a(del_ATPase) | | Descriptor: | IMITATION SWITCH PROTEIN 1 (DEL_ATPASE), ISWI ONE COMPLEX PROTEIN 3 | | Authors: | Yamada, K, Frouws, T.D, Angst, B, Fitzgerald, D.J, DeLuca, C, Schimmele, K, Sargent, D.F, Richmond, T.J. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and Mechanism of the Chromatin Remodelling Factor Isw1A.
Nature, 472, 2011
|
|
2YL8
 
 | | Inhibition of the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, BETA-N-ACETYLHEXOSAMINIDASE, ... | | Authors: | Pluvinage, B, Higgins, M.A, Abbott, D.W, Robb, C, Dalia, A.B, Deng, L, Weiser, J.N, Parsons, T.B, Fairbanks, A.J, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-05-31 | | Release date: | 2011-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inhibition of the Pneumococcal Virulence Factor Strh and Molecular Insights Into N-Glycan Recognition and Hydrolysis.
Structure, 19, 2011
|
|
2YBB
 
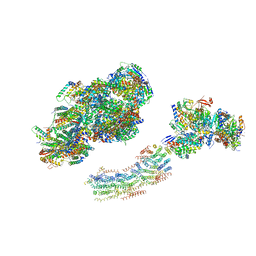 | | Fitted model for bovine mitochondrial supercomplex I1III2IV1 by single particle cryo-EM (EMD-1876) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Althoff, T, Mills, D.J, Popot, J.-L, Kuehlbrandt, W. | | Deposit date: | 2011-03-02 | | Release date: | 2011-10-19 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Arrangement of Electron Transport Chain Components in Bovine Mitochondrial Supercomplex I(1)III(2)Iv(1).
Embo J., 30, 2011
|
|
2Y8I
 
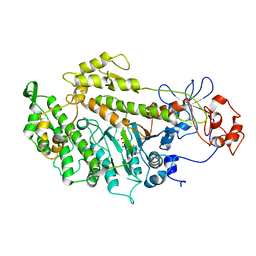 | | Structural basis for the allosteric interference of myosin function by mutants G680A and G680V of Dictyostelium myosin-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN-2 HEAVY CHAIN | | Authors: | Preller, M, Bauer, S, Adamek, N, Fujita-Becker, S, Fedorov, R, Geeves, M.A, Manstein, D.J. | | Deposit date: | 2011-02-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.132 Å) | | Cite: | Structural Basis for the Allosteric Interference of Myosin Function by Reactive Thiol Region Mutations G680A and G680V.
J.Biol.Chem., 286, 2011
|
|
2YL5
 
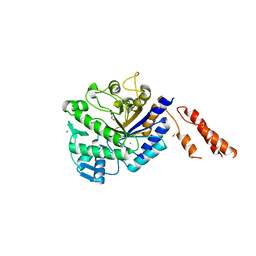 | | Inhibition of the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis | | Descriptor: | 1,2-ETHANEDIOL, BETA-N-ACETYLHEXOSAMINIDASE, MAGNESIUM ION | | Authors: | Pluvinage, B, Higgins, M.A, Abbott, D.W, Robb, C, Dalia, A.B, Deng, L, Weiser, J.N, Parsons, T.B, Fairbanks, A.J, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-05-31 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Inhibition of the Pneumococcal Virulence Factor Strh and Molecular Insights Into N-Glycan Recognition and Hydrolysis.
Structure, 19, 2011
|
|
2YLL
 
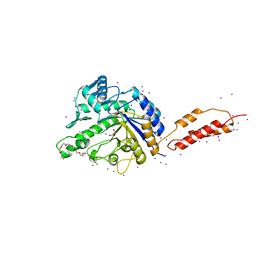 | | Inhibition of the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, BETA-N-ACETYLHEXOSAMINIDASE, ... | | Authors: | Pluvinage, B, Higgins, M.A, Abbott, D.W, Robb, C, Dalia, A.B, Deng, L, Weiser, J.N, Parsons, T.B, Fairbanks, A.J, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-06-02 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Inhibition of the Pneumococcal Virulence Factor Strh and Molecular Insights Into N-Glycan Recognition and Hydrolysis.
Structure, 19, 2011
|
|
2YA2
 
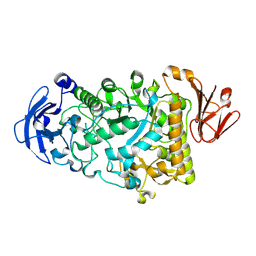 | | Catalytic Module of the Multi-modular glycogen-degrading pneumococcal virulence factor SpuA in complex with an inhibitor. | | Descriptor: | CALCIUM ION, PUTATIVE ALKALINE AMYLOPULLULANASE, SODIUM ION, ... | | Authors: | Lammerts van Bueren, A, Ficko-Blean, E, Pluvinage, B, Hehemann, J.H, Higgins, M.A, Deng, L, Ogunniyi, A.D, Stroeher, U.H, Warry, N.E, Burke, R.D, Czjzek, M, Paton, J.C, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The Conformation and Function of a Multimodular Glycogen-Degrading Pneumococcal Virulence Factor.
Structure, 19, 2011
|
|
2Y73
 
 | | THE NATIVE STRUCTURES OF SOLUBLE HUMAN PRIMARY AMINE OXIDASE AOC3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Elovaara, H, Kidron, H, Parkash, V, Nymalm, Y, Bligt, E, Ollikka, P, Smith, D.J, Pihlavisto, M, Salmi, M, Jalkanen, S, Salminen, T.A. | | Deposit date: | 2011-01-28 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Two Imidazole Binding Sites and Key Residues for Substrate Specificity in Human Primary Amine Oxidase Aoc3.
Biochemistry, 50, 2011
|
|
2Y9Z
 
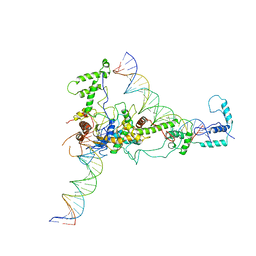 | | Chromatin Remodeling Factor ISW1a(del_ATPase) in DNA complex | | Descriptor: | I-DNA/E-DNA, IMITATION SWITCH PROTEIN 1 (DEL_ATPASE), ISWI ONE COMPLEX PROTEIN 3 | | Authors: | Yamada, K, Frouws, T.D, Angst, B, Fitzgerald, D.J, DeLuca, C, Schimmele, K, Sargent, D.F, Richmond, T.J. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.601 Å) | | Cite: | Structure and Mechanism of the Chromatin Remodelling Factor Isw1A
Nature, 472, 2011
|
|
2Z57
 
 | | Crystal structure of G56E-propeptide:S324A-subtilisin complex | | Descriptor: | CALCIUM ION, Tk-subtilisin, ZINC ION | | Authors: | Pulido, M.A, Tanaka, S, Sringiew, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-06-29 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Requirement of left-handed glycine residue for high stability of the Tk-subtilisin propeptide as revealed by mutational and crystallographic analyses
J.Mol.Biol., 374, 2007
|
|
2Z56
 
 | | Crystal structure of G56S-propeptide:S324A-subtilisin complex | | Descriptor: | CALCIUM ION, Tk-subtilisin, ZINC ION | | Authors: | Pulido, M.A, Tanaka, S, Sringiew, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-06-29 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Requirement of left-handed glycine residue for high stability of the Tk-subtilisin propeptide as revealed by mutational and crystallographic analyses
J.Mol.Biol., 374, 2007
|
|
2Z8Z
 
 | | Crystal structure of a platinum-bound S445C mutant of Pseudomonas sp. MIS38 lipase | | Descriptor: | CALCIUM ION, Lipase, PLATINUM (II) ION, ... | | Authors: | Angkawidjaja, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-09-13 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a family I.3 lipase from Pseudomonas sp. MIS38 in a closed conformation
FEBS Lett., 581, 2007
|
|
2Z58
 
 | | Crystal structure of G56W-propeptide:S324A-subtilisin complex | | Descriptor: | CALCIUM ION, Tk-subtilisin, ZINC ION | | Authors: | Pulido, M.A, Tanaka, S, Sringiew, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-06-29 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Requirement of left-handed glycine residue for high stability of the Tk-subtilisin propeptide as revealed by mutational and crystallographic analyses
J.Mol.Biol., 374, 2007
|
|
7B6W
 
 | | Crystal structure of the human alpha1B adrenergic receptor in complex with inverse agonist (+)-cyclazosin | | Descriptor: | Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor, [(4~{a}~{R},8~{a}~{S})-4-(4-azanyl-6,7-dimethoxy-quinazolin-2-yl)-2,3,4~{a},5,6,7,8,8~{a}-octahydroquinoxalin-1-yl]-(furan-2-yl)methanone | | Authors: | Deluigi, M, Morstein, L, Hilge, M, Schuster, M, Merklinger, L, Klipp, A, Scott, D.J, Plueckthun, A. | | Deposit date: | 2020-12-08 | | Release date: | 2022-01-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.873 Å) | | Cite: | Crystal structure of the alpha 1B -adrenergic receptor reveals molecular determinants of selective ligand recognition.
Nat Commun, 13, 2022
|
|
3AKJ
 
 | |
7BNO
 
 | | Open conformation of D614G SARS-CoV-2 spike with 2 Erect RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BNM
 
 | | Closed conformation of D614G SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3AKK
 
 | |
3AKL
 
 | |
2Y9E
 
 | | Structural basis for the allosteric interference of myosin function by mutants G680A and G680V of Dictyostelium myosin-2 | | Descriptor: | MYOSIN-2 | | Authors: | Preller, M, Bauer, S, Adamek, N, Fujita-Becker, S, Fedorov, R, Geeves, M.A, Manstein, D.J. | | Deposit date: | 2011-02-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.397 Å) | | Cite: | Structural Basis for the Allosteric Interference of Myosin Function by Reactive Thiol Region Mutations G680A and G680V.
J.Biol.Chem., 286, 2011
|
|
7BLR
 
 | | Vps35/Vps29 arch of fungal membrane-assembled retromer:Vps5 (SNX-BAR) complex. | | Descriptor: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Leneva, N, Kovtun, O, Morado, D.R, Briggs, J.A.G, Owen, D.J. | | Deposit date: | 2021-01-18 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly.
Sci Adv, 7, 2021
|
|
7BLP
 
 | | Vps35/Vps29 arch of fungal membrane-assembled retromer:Grd19 complex | | Descriptor: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Leneva, N, Kovtun, O, Morado, D.R, Briggs, J.A.G, Owen, D.J. | | Deposit date: | 2021-01-18 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly.
Sci Adv, 7, 2021
|
|
7BLN
 
 | | VPS35/VPS29 arch of metazoan membrane-assembled retromer:SNX3 complex modelled with human proteins | | Descriptor: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Leneva, N, Kovtun, O, Morado, D.R, Briggs, J.A.G, Owen, D.J. | | Deposit date: | 2021-01-18 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly.
Sci Adv, 7, 2021
|
|
2YA0
 
 | | Catalytic Module of the Multi-modular glycogen-degrading pneumococcal virulence factor SpuA | | Descriptor: | CALCIUM ION, GLYCEROL, PUTATIVE ALKALINE AMYLOPULLULANASE, ... | | Authors: | Lammerts van Bueren, A, Ficko-Blean, E, Pluvinage, B, Hehemann, J.H, Higgins, M.A, Deng, L, Ogunniyi, A.D, Stroeher, U.H, Warry, N.E, Burke, R.D, Czjzek, M, Paton, J.C, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Conformation and Function of a Multimodular Glycogen-Degrading Pneumococcal Virulence Factor.
Structure, 19, 2011
|
|
