7NR2
 
 | | The structure of the SBP TarP_Sse in complex with coumarate | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-02 | | Release date: | 2021-10-06 | | Last modified: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NQG
 
 | | The structure of the SBP TarP_Rhp in complex with 4-hydroxyphenylacetate | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYPHENYLACETATE, PHOSPHATE ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NRA
 
 | | The structure of the SBP TarP_Sse in complex with cinnamate | | Descriptor: | HYDROCINNAMIC ACID, TRAP dicarboxylate transporter, DctP subunit | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NSW
 
 | | The structure of the SBP TarP_Csal in complex with coumarate | | Descriptor: | 1,2-ETHANEDIOL, 4'-HYDROXYCINNAMIC ACID, MAGNESIUM ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-08 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NTD
 
 | | The structure of the SBP TarP_Csal in complex with ferulate | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NTE
 
 | | The structure of an open conformation of the SBP TarP_Csal | | Descriptor: | MAGNESIUM ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NXF
 
 | | Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - monomer unit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Heit, S, Geurts, M.M.G, Murphy, B.J, Corey, R, Mills, D.J, Kuehlbrandt, W, Bublitz, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the hexameric fungal plasma membrane proton pump in its autoinhibited state.
Sci Adv, 7, 2021
|
|
7OBM
 
 | | Crystal structure of the human Prolyl Endopeptidase-Like protein short form (residues 90-727) | | Descriptor: | Prolyl endopeptidase-like | | Authors: | Rosier, K, McDevitt, M.T, Brendan, J.F, Marcaida, M.J, Bingman, C.A, Pagliarini, D.J, Creemers, J.W.M, Smith, R.W, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Prolyl endopeptidase-like is a (thio)esterase involved in mitochondrial respiratory chain function.
Iscience, 24, 2021
|
|
7NY1
 
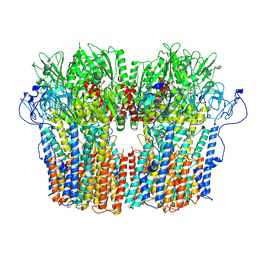 | | Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - hexameric assembly | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Heit, S, Geurts, M.M.G, Murphy, B.J, Corey, R, Mills, D.J, Kuehlbrandt, W, Bublitz, M. | | Deposit date: | 2021-03-19 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the hexameric fungal plasma membrane proton pump in its autoinhibited state.
Sci Adv, 7, 2021
|
|
8UPV
 
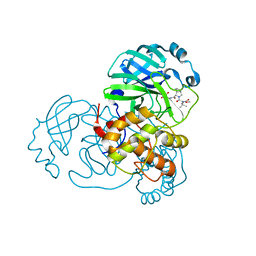 | | Structure of SARS-Cov2 3CLPro in complex with Compound 33 | | Descriptor: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6R)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UPS
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 5 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, PHOSPHATE ION | | Authors: | Wu, Y, Qiang, D, Zhuang, N, Krishnamurthy, H, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UAU
 
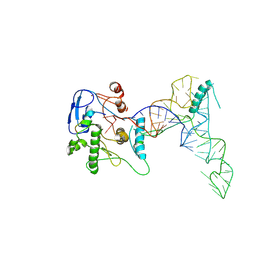 | | human ATE1 in complex with Arg-tRNA and a peptide substrate | | Descriptor: | Isoform ATE1-2 of Arginyl-tRNA--protein transferase 1, RNA (76-MER), ZINC ION, ... | | Authors: | Huang, W, Zhang, Y, Taylor, D.J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Oligomerization and a distinct tRNA-binding loop are important regulators of human arginyl-transferase function.
Nat Commun, 15, 2024
|
|
8URQ
 
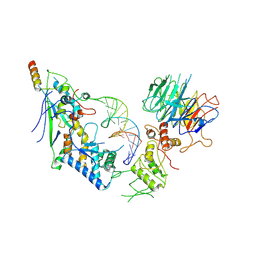 | | Spo11 core complex with gapped DNA | | Descriptor: | Antiviral protein SKI8, MAGNESIUM ION, Meiosis-specific protein SPO11, ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the Spo11 core complex bound to DNA.
Nat.Struct.Mol.Biol., 2024
|
|
8V1J
 
 | |
8V14
 
 | | Structure of NRAP-1 and its role in NMDAR signaling | | Descriptor: | CALCIUM ION, NMDA receptor auxiliary protein | | Authors: | Whitby, F.G, Goodell, D.J, Maricq, A.V, Hill, C.P. | | Deposit date: | 2023-11-19 | | Release date: | 2024-01-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic and structural studies reveal NRAP-1-dependent coincident activation of NMDARs.
Cell Rep, 43, 2024
|
|
7BC4
 
 | | Cryo-EM structure of fatty acid synthase (FAS) from Pichia pastoris | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | Authors: | Snowden, J.S, Alzahrani, J, Sherry, L, Stacey, M, Rowlands, D.J, Ranson, N.A, Stonehouse, N.J. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into Pichia pastoris fatty acid synthase.
Sci Rep, 11, 2021
|
|
7BC5
 
 | | Cryo-EM structure of ACP domain from Pichia pastoris fatty acid synthase (FAS) | | Descriptor: | Fatty acid synthase subunit alpha | | Authors: | Snowden, J.S, Alzahrani, J, Sherry, L, Stacey, M, Rowlands, D.J, Ranson, N.A, Stonehouse, N.J. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into Pichia pastoris fatty acid synthase.
Sci Rep, 11, 2021
|
|
4W6C
 
 | |
4W6F
 
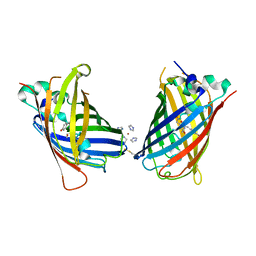 | | Crystal Structure of Full-Length Split GFP Mutant K26C Disulfide Dimer, P 32 2 1 Space Group, Form 2 | | Descriptor: | IMIDAZOLE, NICKEL (II) ION, fluorescent protein D21H/K26C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W6K
 
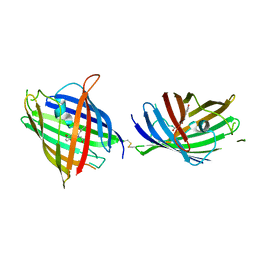 | |
4W74
 
 | |
7BCQ
 
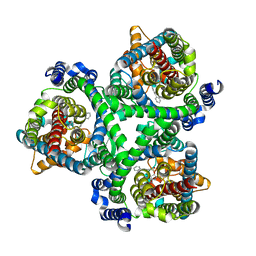 | | ASCT2 in the presence of the inhibitor Lc-BPE (position "up") in the outward-open conformation. | | Descriptor: | 4-(4-phenylphenyl)carbonyloxypyrrolidine-2-carboxylic acid, Neutral amino acid transporter B(0) | | Authors: | Garibsingh, R.A, Ndaru, E, Garaeva, A.A, Shi, Y, Zielewicz, L, Bonomi, M, Slotboom, D.J, Paulino, C, Grewer, C, Schlessinger, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Rational design of ASCT2 inhibitors using an integrated experimental-computational approach.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BCT
 
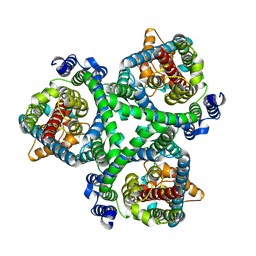 | | ASCT2 in the presence of the inhibitor ERA-21 in the outward-open conformation. | | Descriptor: | Neutral amino acid transporter B(0) | | Authors: | Garibsingh, R.A, Ndaru, E, Garaeva, A.A, Shi, Y, Zielewicz, L, Bonomi, M, Slotboom, D.J, Paulino, C, Grewer, C, Schlessinger, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Rational design of ASCT2 inhibitors using an integrated experimental-computational approach.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BMK
 
 | | ATP-Competitive Partial Antagonists-'PAIR's-Rheostatically Modulate IRE1alpha's Kinase Helix-alphaC to Segregate its RNase-Mediated Biological Outputs | | Descriptor: | 1,2-ETHANEDIOL, 2,2,2-tris(fluoranyl)-~{N}-[4-[3-[2-[[(3~{S})-piperidin-3-yl]amino]pyrimidin-4-yl]pyridin-2-yl]oxynaphthalen-1-yl]ethanesulfonamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feldman, H.C, Ghosh, R, Auyeung, V, Mueller, J.L, Vidadala, V.N, Olivier, A, Backes, B.J, Zikherman, J, Papa, F.R, Maly, D.J. | | Deposit date: | 2021-01-20 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | ATP-competitive partial antagonists of the IRE1 alpha RNase segregate outputs of the UPR.
Nat.Chem.Biol., 17, 2021
|
|
4W7F
 
 | |
