4QEO
 
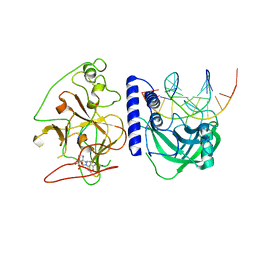 | | crystal structure of KRYPTONITE in complex with mCHH DNA, H3(1-15) peptide and SAH | | Descriptor: | DNA 5'-ACTGATGAGTACCAT-3', DNA 5'-GGTACT(5CM)ATCAGTAT-3', Histone H3, ... | | Authors: | Du, J, Li, S, Patel, D.J. | | Deposit date: | 2014-05-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
4TTM
 
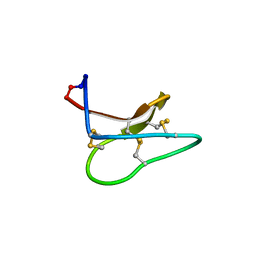 | | Racemic structure of kalata B1 (kB1) | | Descriptor: | D-kalata B1, Kalata-B1 | | Authors: | Wang, C.K, King, G.J, Craik, D.J. | | Deposit date: | 2014-06-22 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9001 Å) | | Cite: | Racemic and Quasi-Racemic X-ray Structures of Cyclic Disulfide-Rich Peptide Drug Scaffolds.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4RSD
 
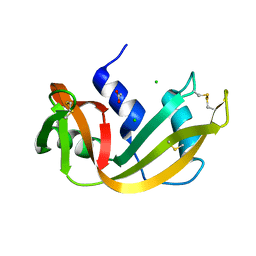 | | STRUCTURE OF THE D121A VARIANT OF RIBONUCLEASE A | | Descriptor: | ACETATE ION, CHLORIDE ION, RIBONUCLEASE A | | Authors: | Schultz, L.W, Quirk, D.J, Raines, R.T. | | Deposit date: | 1998-02-05 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | His...Asp catalytic dyad of ribonuclease A: structure and function of the wild-type, D121N, and D121A enzymes.
Biochemistry, 37, 1998
|
|
2KUK
 
 | |
2KYO
 
 | |
2KPR
 
 | |
2OQF
 
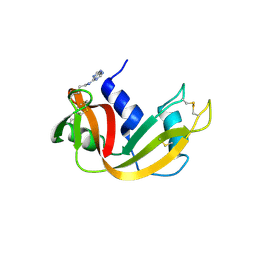 | | Structure of a synthetic, non-natural analogue of RNase A: [N71K(Ade), D83A]RNase A | | Descriptor: | Ribonuclease pancreatic | | Authors: | Boerema, D.J, Tereshko, V.A, Zhang, J.L, He, C, Kent, S.B.H. | | Deposit date: | 2007-01-31 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, Synthesis, and Characterization of Non-natural RNase A Analogues with Enhanced Second-step Catalytic Activity
To be Published
|
|
2OZY
 
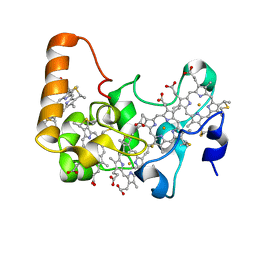 | | Crystal structure of E.coli nrfB | | Descriptor: | Cytochrome c-type protein nrfB, HEME C | | Authors: | Clarke, T.A, Richardson, D.J, Hemmings, A.M. | | Deposit date: | 2007-02-28 | | Release date: | 2007-08-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The crystal structure of the pentahaem c-type cytochrome NrfB and characterization of its solution-state interaction with the pentahaem nitrite reductase NrfA.
Biochem.J., 406, 2007
|
|
2ORX
 
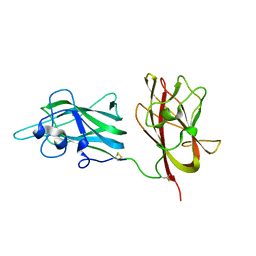 | | Structural Basis for Ligand Binding and Heparin Mediated Activation of Neuropilin | | Descriptor: | Neuropilin-1 | | Authors: | Vander Kooi, C.W, Jusino, M.A, Perman, B, Neau, D.B, Bellamy, H.D, Leahy, D.J. | | Deposit date: | 2007-02-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ligand and heparin binding to neuropilin B domains
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2M2Y
 
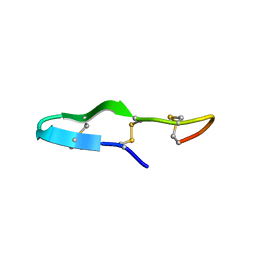 | | Solution structure of the antimicrobial peptide Btd-2[3,4] | | Descriptor: | BTD-2[3,4] | | Authors: | Conibear, A.C, Rosengren, K, Daly, N.L, Troiera Henriques, S, Craik, D.J. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The cyclic cystine ladder in theta-defensins is important for structure and stability, but not antibacterial activity.
J.Biol.Chem., 288, 2013
|
|
2MN1
 
 | | Solution Structure of kalata B1[W23WW] | | Descriptor: | kalata B1[W23WW] | | Authors: | Henriques, S.T, Huang, Y.H, Chaousis, S, Wang, C.K, Craik, D.J. | | Deposit date: | 2014-03-26 | | Release date: | 2015-05-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Anticancer and toxic properties of cyclotides are dependent on phosphatidylethanolamine phospholipid targeting.
Chembiochem, 15, 2014
|
|
2M13
 
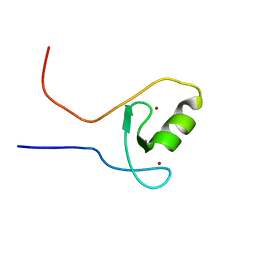 | | The ZZ domain of cytoplasmic polyadenylation element binding protein 1 (CPEB1) | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 1, ZINC ION | | Authors: | Lee, B.M, Merkel, D.J, Wells, S.B, Hilburn, B.C, Elazzouzi, F, Perez-Alvarado, G.C. | | Deposit date: | 2012-11-14 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The C-Terminal Region of Cytoplasmic Polyadenylation Element Binding Protein Is a ZZ Domain with Potential for Protein-Protein Interactions.
J.Mol.Biol., 425, 2013
|
|
2M77
 
 | | [Asp2]RTD-1 | | Descriptor: | [Asp2]RTD-1 | | Authors: | Conibear, A.C, Bochen, A, Rosengren, K, Kessler, H, Craik, D.J. | | Deposit date: | 2013-04-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Cyclic Cystine Ladder of Theta-Defensins as a Stable, Bifunctional Scaffold: A Proof-of-Concept Study Using the Integrin-Binding RGD Motif
Chembiochem, 15, 2014
|
|
2MFA
 
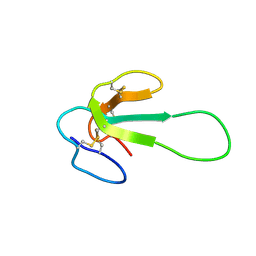 | | Mambalgin-2 | | Descriptor: | Mambalgin-2 | | Authors: | Schroeder, C.I, Rash, L.D, Vila-Farres, X, Rosengren, K.J, Mobli, M, King, G.F, Alewood, P.F, Craik, D.J, Durek, T. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Chemical synthesis, 3D structure, and ASIC binding site of the toxin mambalgin-2.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2LYF
 
 | |
2M98
 
 | |
2LYE
 
 | | High resolution NMR solution structure of a symmetrical theta-defensin, BTD-2 | | Descriptor: | BTD-2 | | Authors: | Conibear, A.C, Rosengren, K, Harvey, P.J, Craik, D.J. | | Deposit date: | 2012-09-18 | | Release date: | 2012-11-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the cyclic cystine ladder motif of theta-defensins.
Biochemistry, 51, 2012
|
|
2P8Z
 
 | | Fitted structure of ADPR-eEF2 in the 80S:ADPR-eEF2:GDPNP:sordarin cryo-EM reconstruction | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, Elongation factor Tu-B, ... | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
2KGG
 
 | |
2KHB
 
 | |
2K7O
 
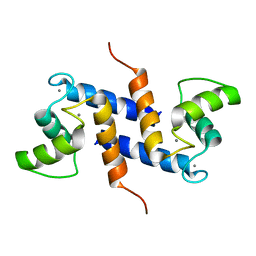 | | Ca2+-S100B, refined with RDCs | | Descriptor: | CALCIUM ION, Protein S100-B | | Authors: | Wright, N.T, Inman, K.G, Levine, J.A, Weber, D.J. | | Deposit date: | 2008-08-17 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refinement of the solution structure and dynamic properties of Ca(2+)-bound rat S100B.
J.Biomol.Nmr, 42, 2008
|
|
2KF3
 
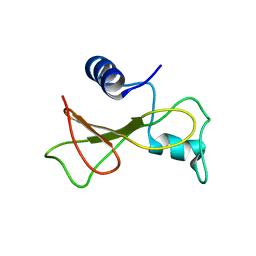 | |
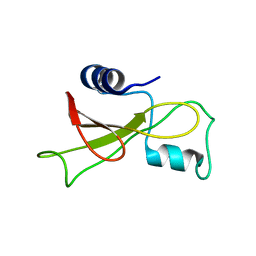
2KF5
 
 | |
2P8W
 
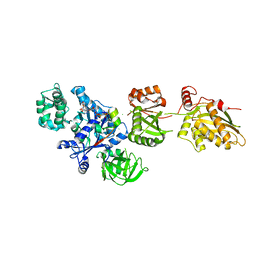 | | Fitted structure of eEF2 in the 80S:eEF2:GDPNP cryo-EM reconstruction | | Descriptor: | Elongation factor 2, Elongation factor Tu-B, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
2OXM
 
 | | Crystal structure of a UNG2/modified DNA complex that represent a stabilized short-lived extrahelical state in ezymatic DNA base flipping | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*(4MF)P*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*AP*TP*CP*TP*T)-3'), Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Stivers, J.T, Amzel, L.M. | | Deposit date: | 2007-02-20 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzymatic capture of an extrahelical thymine in the search for uracil in DNA.
Nature, 449, 2007
|
|
