2VU3
 
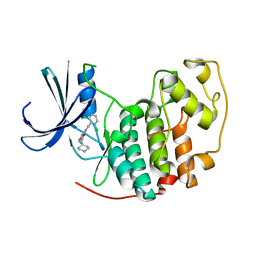 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | 4-{[(2,6-dichlorophenyl)carbonyl]amino}-N-piperidin-4-yl-1H-pyrazole-3-carboxamide, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-20 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
3RR3
 
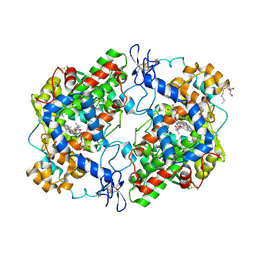 | | Structure of (R)-flurbiprofen bound to mCOX-2 | | Descriptor: | (2R)-2-(3-fluoro-4-phenyl-phenyl)propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Duggan, K.C, Hermanson, D.J, Musee, J, Prusakiewicz, J.J, Scheib, J, Carter, B.D, Banerjee, S, Oates, J.A, Marnett, L.J. | | Deposit date: | 2011-04-28 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.842 Å) | | Cite: | (R)-Profens are substrate-selective inhibitors of endocannabinoid oxygenation by COX-2.
Nat.Chem.Biol., 7, 2011
|
|
3RV1
 
 | | Crystal structure of the N-terminal and RNase III domains of K. polysporus Dcr1 E224Q mutant | | Descriptor: | K. polysporus Dcr1 | | Authors: | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | Deposit date: | 2011-05-05 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | The inside-out mechanism of dicers from budding yeasts.
Cell(Cambridge,Mass.), 146, 2011
|
|
3RYO
 
 | | Crystal Structure of Enhanced Intracellular Survival (Eis) Protein from Mycobacterium tuberculosis with Acetyl CoA | | Descriptor: | ACETYL COENZYME *A, Enhanced intracellular survival protein | | Authors: | Kim, K.H, An, D.R, Yoon, J.Y, Kim, H.S, Yoon, H.J, Song, J, Im, H.N, Kim, J, Kim, D.J, Lee, S.J, Kim, H.J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2011-05-11 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7
Proc.Natl.Acad.Sci.USA, 2012
|
|
3RVH
 
 | | Crystal Structure of JMJD2A Complexed with Inhibitor | | Descriptor: | 8-hydroxy-3-(piperazin-1-yl)quinoline-5-carboxylic acid, GLYCEROL, Lysine-specific demethylase 4A, ... | | Authors: | King, O.N.F, Maloney, D.J, Tumber, A, Rai, G, Jadhav, A, Clifton, I.J, Heightman, T.D, Simeonov, A, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Crystal Structure of JMJD2A Complexed with Inhibitor
To be Published
|
|
2W92
 
 | | Structure of a Streptococcus pneumoniae family 85 glycoside hydrolase, Endo-D, in complex with NAG-thiazoline. | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ENDO-BETA-N-ACETYLGLUCOSAMINIDASE D, PENTAETHYLENE GLYCOL | | Authors: | Abbott, D.W, Macauley, M.S, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2009-01-21 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Streptococcus Pneumoniae Endohexosaminidase D, Structural and Mechanistic Insight Into Substrate-Assisted Catalysis in Family 85 Glycoside Hydrolases.
J.Biol.Chem., 284, 2009
|
|
2W8U
 
 | | SPT with PLP, N100Y | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE PALMITOYLTRANSFERASE | | Authors: | Raman, M.C.C, Johnson, K.A, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2009-01-19 | | Release date: | 2009-01-27 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The External-Aldimine Form of Serine Palmitoyltranserase; Structural, Kinetic and Spectroscopic Analysis of the Wild-Type Enzyme and Hsan1 Mutant Mimics.
J.Biol.Chem., 284, 2009
|
|
3RW6
 
 | | Structure of nuclear RNA export factor TAP bound to CTE RNA | | Descriptor: | Nuclear RNA export factor 1, constitutive transport element(CTE)of Mason-Pfizer monkey virus RNA | | Authors: | Teplova, M, Khin, N.W, Patel, D.J, Izaurralde, E. | | Deposit date: | 2011-05-07 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-function studies of nucleocytoplasmic transport of retroviral genomic RNA by mRNA export factor TAP.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2WFO
 
 | | Crystal structure of Machupo virus envelope glycoprotein GP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCOPROTEIN 1 | | Authors: | Bowden, T.A, Crispin, M, Graham, S.C, Harvey, D.J, Grimes, J.M, Jones, E.Y, Stuart, D.I. | | Deposit date: | 2009-04-09 | | Release date: | 2009-06-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Unusual Molecular Architecture of the Machupo Virus Attachment Glycoprotein.
J.Virol., 83, 2009
|
|
3RWV
 
 | | Crystal Structure of apo-form of Human Glycolipid Transfer Protein at 1.5 A resolution | | Descriptor: | Glycolipid transfer protein, SULFATE ION | | Authors: | Samygina, V, Cabo-Bilbao, A, Popov, A.N, Ochoa-Lizarralde, B, Goni-de-Cerio, F, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-09 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3S0K
 
 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with glucosylceramide containing oleoyl acyl chain (18:1) | | Descriptor: | (9Z)-N-[(2S,3R,4E)-1-(beta-D-glucopyranosyloxy)-3-hydroxyoctadec-4-en-2-yl]octadec-9-enamide, Glycolipid transfer protein, NICKEL (II) ION | | Authors: | Cabo-Bilbao, A, Samygina, V, Popov, A.N, Ochoa-Lizarralde, B, Goni-de-Cerio, F, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-13 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3RW7
 
 | | Structure of N-terminal domain of nuclear RNA export factor TAP | | Descriptor: | Nuclear RNA export factor 1 | | Authors: | Teplova, M, Khin, N.W, Wohlbold, L, Izaurralde, E, Patel, D.J. | | Deposit date: | 2011-05-07 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-function studies of nucleocytoplasmic transport of retroviral genomic RNA by mRNA export factor TAP.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3S0I
 
 | | Crystal Structure of D48V mutant of Human Glycolipid Transfer Protein complexed with 3-O-sulfo galactosylceramide containing nervonoyl acyl chain | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Popov, A.N, Cabo-Bilbao, A, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-13 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
4LOH
 
 | | Crystal structure of hSTING(H232) in complex with c[G(2',5')pA(3',5')p] | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
4LOJ
 
 | | Crystal structure of mSting in complex with c[G(2',5')pA(3',5')p] | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
8UPW
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 34 | | Descriptor: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6S)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UTE
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 27 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S)-6,6-difluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-31 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8U4W
 
 | |
7O9J
 
 | | Crystal structure of DyP-type peroxidase from Dictyostelium discoideum in complex with an activated form of oxygen | | Descriptor: | 1,2-ETHANEDIOL, DyPA, OXYGEN MOLECULE, ... | | Authors: | Rai, A, Fedorov, R, Manstein, D.J. | | Deposit date: | 2021-04-16 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Characterization of a Dye-Decolorizing Peroxidase from Dictyostelium discoideum .
Int J Mol Sci, 22, 2021
|
|
7O9L
 
 | | Dictyostelium discoideum dye decolorizing peroxidase DyPA in complex with cyanide. | | Descriptor: | 1,2-ETHANEDIOL, CYANIDE ION, DyPA, ... | | Authors: | Rai, A, Fedorov, R, Manstein, D.J. | | Deposit date: | 2021-04-16 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Characterization of a Dye-Decolorizing Peroxidase from Dictyostelium discoideum .
Int J Mol Sci, 22, 2021
|
|
7ODZ
 
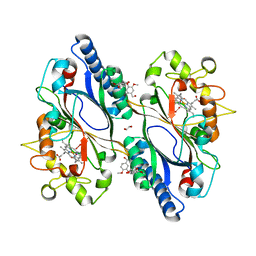 | | Dictyostelium discoideum dye decolorizing peroxidase DyPA in complex with an activated form of oxygen and veratryl alcohol | | Descriptor: | 1,2-ETHANEDIOL, DyPA, OXYGEN MOLECULE, ... | | Authors: | Rai, A, Fedorov, R, Manstein, D.J. | | Deposit date: | 2021-04-30 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Characterization of a Dye-Decolorizing Peroxidase from Dictyostelium discoideum .
Int J Mol Sci, 22, 2021
|
|
7NNU
 
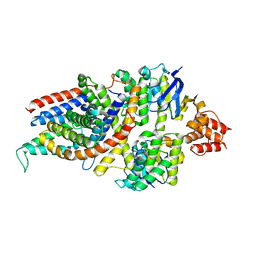 | | Cryo-EM structure of the folate-specific ECF transporter complex in MSP2N2 lipid nanodiscs | | Descriptor: | Conserved hypothetical membrane protein, Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2021-02-25 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Insights into the bilayer-mediated toppling mechanism of a folate-specific ECF transporter by cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NNT
 
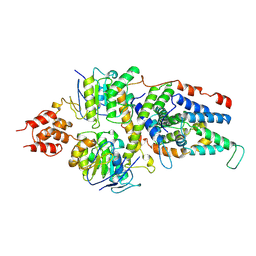 | | Cryo-EM structure of the folate-specific ECF transporter complex in DDM micelles | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2021-02-25 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Insights into the bilayer-mediated toppling mechanism of a folate-specific ECF transporter by cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8URU
 
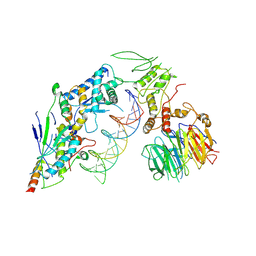 | | Spo11 core complex with hairpin DNA | | Descriptor: | Antiviral protein SKI8, Hairpin DNA, MAGNESIUM ION, ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of the Spo11 core complex bound to DNA.
Nat.Struct.Mol.Biol., 2024
|
|
7NRR
 
 | | The structure of the SBP TarP_Csal in complex with caffeate | | Descriptor: | CAFFEIC ACID, MAGNESIUM ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-04 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
