4KFO
 
 | | Structure-Based Discovery of Novel Amide-Containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}imidazo[1,2-a]pyridine-6-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Gunzner-Tosteb, J, Liederer, B.M, Ly, J, O'Brien, T, Oh, A, Wang, L, Wang, W, Xiao, Y, Zak, M, Zhao, G, Yuen, P, Bair, K.W. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
4NH3
 
 | |
4NT2
 
 | | Crystal structure of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with lyso-sphingomyelin (d18:1) at 2.4 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(R)-{[(2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl]oxy}(hydroxy)phosphoryl]oxy}-N,N,N-trimethylethanaminium, SULFATE ION, ... | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
4NTI
 
 | | Crystal structure of D60N mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C12 ceramide-1-phosphate (d18:1/12:0) at 2.9 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, DI(HYDROXYETHYL)ETHER, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
3U6B
 
 | | Ef-tu (escherichia coli) in complex with nvp-ldi028 | | Descriptor: | Elongation factor Tu 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Palestrant, D.J. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Antibacterial optimization of 4-aminothiazolyl analogues of the natural product GE2270 A: identification of the cycloalkylcarboxylic acids.
J.Med.Chem., 54, 2011
|
|
1DVN
 
 | |
1DF6
 
 | |
1DJ9
 
 | | CRYSTAL STRUCTURE OF 8-AMINO-7-OXONANOATE SYNTHASE (OR 7-KETO-8AMINIPELARGONATE OR KAPA SYNTHASE) COMPLEXED WITH PLP AND THE PRODUCT 8(S)-AMINO-7-OXONANONOATE (OR KAPA). THE ENZYME OF BIOTIN BIOSYNTHETIC PATHWAY. | | Descriptor: | 8-AMINO-7-OXONONANOATE SYNTHASE, MAGNESIUM ION, N-[7-KETO-8-AMINOPELARGONIC ACID]-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Webster, S.P, Alexeev, D, Campopiano, D.J, Watt, R.M, Alexeeva, M, Sawyer, L, Baxter, R.L. | | Deposit date: | 1999-12-02 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of 8-amino-7-oxononanoate synthase: spectroscopic, kinetic, and crystallographic studies.
Biochemistry, 39, 2000
|
|
1EVH
 
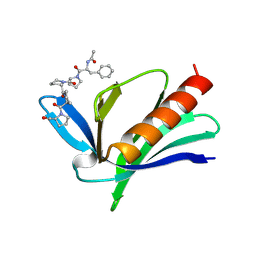 | |
1EXV
 
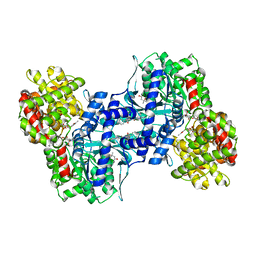 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE A COMPLEXED WITH GLCNAC AND CP-403,700 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, LIVER GLYCOGEN PHOSPHORYLASE, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Rath, V.L, Ammirati, M, Danley, D.E, Ekstrom, J.L, Hynes, T.R, Olson, T.V, Hoover, D.J. | | Deposit date: | 2000-05-04 | | Release date: | 2000-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human liver glycogen phosphorylase inhibitors bind at a new allosteric site.
Chem.Biol., 7, 2000
|
|
1EXY
 
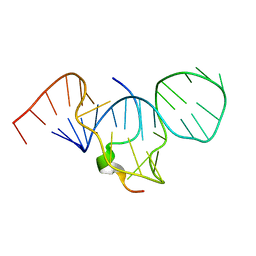 | | SOLUTION STRUCTURE OF HTLV-1 PEPTIDE BOUND TO ITS RNA APTAMER TARGET | | Descriptor: | HTLV-1 REX PEPTIDE, RNA APTAMER, 33-MER | | Authors: | Jiang, F, Gorin, A, Hu, W, Majumdar, A, Baskerville, S, Xu, W, Ellington, A, Patel, D.J. | | Deposit date: | 2000-05-05 | | Release date: | 2000-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Anchoring an extended HTLV-1 Rex peptide within an RNA major groove containing junctional base triples.
Structure Fold.Des., 7, 1999
|
|
1F40
 
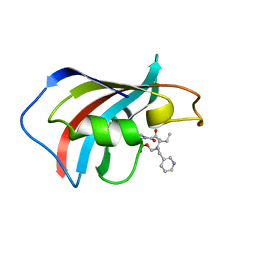 | | SOLUTION STRUCTURE OF FKBP12 COMPLEXED WITH GPI-1046, A NEUROTROPHIC LIGAND | | Descriptor: | (2S)-[3-PYRIDYL-1-PROPYL]-1-[3,3-DIMETHYL-1,2-DIOXOPENTYL]-2-PYRROLIDINECARBOXYLATE, FK506 BINDING PROTEIN (FKBP12) | | Authors: | Sich, C, Improta, S, Cowley, D.J, Guenet, C, Merly, J.P, Teufel, M, Saudek, V. | | Deposit date: | 2000-06-07 | | Release date: | 2000-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a neurotrophic ligand bound to FKBP12 and its effects on protein dynamics.
Eur.J.Biochem., 267, 2000
|
|
1F4U
 
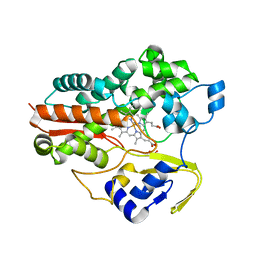 | | THERMOPHILIC P450: CYP119 FROM SULFOLOBUS SOLFACTARICUS | | Descriptor: | CYTOCHROME P450 119, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yano, J.K, Koo, L.S, Schuller, D.J, Li, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2000-06-09 | | Release date: | 2000-10-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of a thermophilic cytochrome P450 from the archaeon Sulfolobus solfataricus.
J.Biol.Chem., 275, 2000
|
|
1G6Q
 
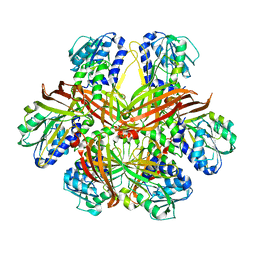 | | CRYSTAL STRUCTURE OF YEAST ARGININE METHYLTRANSFERASE, HMT1 | | Descriptor: | HNRNP ARGININE N-METHYLTRANSFERASE | | Authors: | Weiss, V.H, McBride, A.E, Soriano, M.A, Filman, D.J, Silver, P.A, Hogle, J.M. | | Deposit date: | 2000-11-07 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure and oligomerization of the yeast arginine methyltransferase, Hmt1.
Nat.Struct.Biol., 7, 2000
|
|
1FYB
 
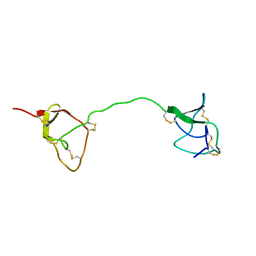 | | SOLUTION STRUCTURE OF C1-T1, A TWO-DOMAIN PROTEINASE INHIBITOR DERIVED FROM THE CIRCULAR PRECURSOR PROTEIN NA-PROPI FROM NICOTIANA ALATA | | Descriptor: | PROTEINASE INHIBITOR | | Authors: | Craik, D.J, Schirra, H.J, Scanlon, M.J, Anderson, M.A. | | Deposit date: | 2000-09-28 | | Release date: | 2001-02-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of C1-T1, a two-domain proteinase inhibitor derived from a circular precursor protein from Nicotiana alata.
J.Mol.Biol., 306, 2001
|
|
1DTO
 
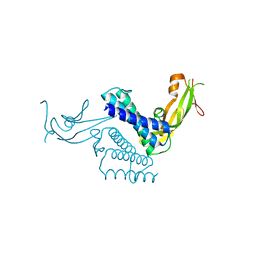 | | CRYSTAL STRUCTURE OF THE COMPLETE TRANSACTIVATION DOMAIN OF E2 PROTEIN FROM THE HUMAN PAPILLOMAVIRUS TYPE 16 | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Antson, A.A, Burns, J.E, Moroz, O.V, Scott, D.J, Sanders, C.M, Bronstein, I.B, Dodson, G.G, Wilson, K.S, Maitland, N. | | Deposit date: | 2000-01-13 | | Release date: | 2000-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the intact transactivation domain of the human papillomavirus E2 protein.
Nature, 403, 2000
|
|
1DVM
 
 | | ACTIVE FORM OF HUMAN PAI-1 | | Descriptor: | CHLORIDE ION, PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Stout, T.J, Graham, H, Buckley, D.I, Matthews, D.J. | | Deposit date: | 2000-01-21 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of active and latent PAI-1: a possible stabilizing role for chloride ions.
Biochemistry, 39, 2000
|
|
1DPS
 
 | | THE CRYSTAL STRUCTURE OF DPS, A FERRITIN HOMOLOG THAT BINDS AND PROTECTS DNA | | Descriptor: | DPS, SODIUM ION | | Authors: | Grant, R.A, Filman, D.J, Finkel, S.E, Kolter, R, Hogle, J.M. | | Deposit date: | 1998-02-23 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Dps, a ferritin homolog that binds and protects DNA.
Nat.Struct.Biol., 5, 1998
|
|
1DSV
 
 | | STRUCTURE OF THE MMTV NUCLEOCAPSID PROTEIN (C-TERMINAL ZINC FINGER) | | Descriptor: | NUCLEIC ACID BINDING PROTEIN P14, ZINC ION | | Authors: | Klein, D.J, Johnson, P.E, Zollars, E.S, De Guzman, R.N, Summers, M.F. | | Deposit date: | 2000-01-08 | | Release date: | 2000-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle.
Biochemistry, 39, 2000
|
|
1DSQ
 
 | | STRUCTURE OF THE MMTV NUCLEOCAPSID PROTEIN (ZINC FINGER 1) | | Descriptor: | NUCLEIC ACID BINDING PROTEIN P14, ZINC ION | | Authors: | Klein, D.J, Johnson, P.E, Zollars, E.S, De Guzman, R.N, Summers, M.F. | | Deposit date: | 2000-01-08 | | Release date: | 2000-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle.
Biochemistry, 39, 2000
|
|
1FG9
 
 | | 3:1 COMPLEX OF INTERFERON-GAMMA RECEPTOR WITH INTERFERON-GAMMA DIMER | | Descriptor: | INTERFERON GAMMA, INTERFERON-GAMMA RECEPTOR ALPHA CHAIN | | Authors: | Thiel, D.J, le Du, M.-H, Walter, R.L, D'Arcy, A, Chene, C, Fountoulakis, M, Garotta, G, Winkler, F.K, Ealick, S.E. | | Deposit date: | 2000-07-28 | | Release date: | 2000-08-11 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Observation of an unexpected third receptor molecule in the crystal structure of human interferon-gamma receptor complex.
Structure Fold.Des., 8, 2000
|
|
1E42
 
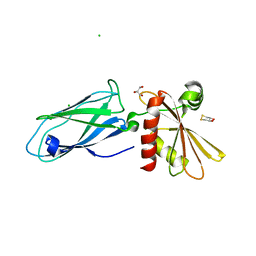 | | Beta2-adaptin appendage domain, from clathrin adaptor AP2 | | Descriptor: | AP-2 COMPLEX SUBUNIT BETA, CHLORIDE ION, DITHIANE DIOL, ... | | Authors: | Owen, D.J, Evans, P.R, McMahon, H.T. | | Deposit date: | 2000-06-27 | | Release date: | 2000-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure and Function of the Beta2-Adaptin Appendage Domain
Embo J., 19, 2000
|
|
1FC0
 
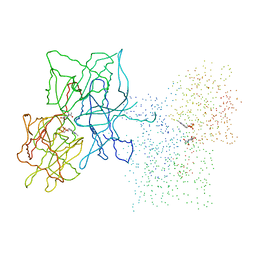 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE COMPLEXED WITH N-ACETYL-BETA-D-GLUCOPYRANOSYLAMINE | | Descriptor: | GLYCOGEN PHOSPHORYLASE, LIVER FORM, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Rath, V.L, Ammirati, M, LeMotte, P.K, Fennell, K.F, Mansour, M.M, Danley, D.E, Hynes, T.R, Schulte, G.K, Wasilko, D.J, Pandit, J. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation of human liver glycogen phosphorylase by alteration of the secondary structure and packing of the catalytic core.
Mol.Cell, 6, 2000
|
|
1FJB
 
 | | NMR Study of an 11-Mer DNA Duplex Containing 7,8-Dihydro-8-Oxoadenine (AOXO) Opposite Thymine | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*(A38)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3') | | Authors: | Chen, H, Johnson, F, Grollman, A.P, Patel, D.J. | | Deposit date: | 1995-12-15 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Studies of the Ionizing Radiation Adduct 7,8-Dihydro-8-Oxoadenine (A Oxo) Positioned Opposite Thymine and Guanine in DNA Duplexes
MAGN.RESON.CHEM., 34, 1996
|
|
1FMN
 
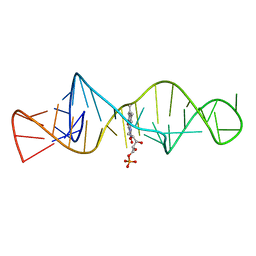 | | SOLUTION STRUCTURE OF FMN-RNA APTAMER COMPLEX, NMR, 5 STRUCTURES | | Descriptor: | FLAVIN MONONUCLEOTIDE, RNA (5'-R(*GP*GP*CP*GP*UP*GP*UP*AP*GP*GP *AP*UP*AP*UP*GP*CP*UP*UP*CP*GP*GP*CP*AP*GP*AP*AP*GP *GP*AP*CP*AP*CP*GP*CP*C)-3') | | Authors: | Fan, P, Suri, A.K, Fiala, R, Live, D, Patel, D.J. | | Deposit date: | 1995-12-04 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular recognition in the FMN-RNA aptamer complex.
J.Mol.Biol., 258, 1996
|
|
