4MDT
 
 | | Structure of LpxC bound to the reaction product UDP-(3-O-(R-3-hydroxymyristoyl))-glucosamine | | Descriptor: | PHOSPHATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION, ... | | Authors: | Clayton, G.M, Klein, D.J, Rickert, K.W, Patel, S.B, Kornienko, M, Zugay-Murphy, J, Reid, J.C, Tummala, S, Sharma, S, Singh, S.B, Miesel, L, Lumb, K.J, Soisson, S.M. | | Deposit date: | 2013-08-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of the Bacterial Deacetylase LpxC Bound to the Nucleotide Reaction Product Reveals Mechanisms of Oxyanion Stabilization and Proton Transfer.
J.Biol.Chem., 288, 2013
|
|
4MEL
 
 | | Crystal Structure of the human USP11 DUSP-UBL domains | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 11 | | Authors: | Harper, S, Gratton, H.E, Cornaciu, I, Oberer, M, Scott, D.J, Emsley, J, Dreveny, I. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structure and Catalytic Regulatory Function of Ubiquitin Specific Protease 11 N-Terminal and Ubiquitin-like Domains.
Biochemistry, 53, 2014
|
|
1F4J
 
 | | STRUCTURE OF TETRAGONAL CRYSTALS OF HUMAN ERYTHROCYTE CATALASE | | Descriptor: | CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Safo, M.K, Musayev, F.N, Wu, S.H, Abraham, D.J, Ko, T.P. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of tetragonal crystals of human erythrocyte catalase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FQ4
 
 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN HYDROXYETHYLENE INHIBITOR CP-108,420 AND YEAST ASPARTIC PROTEINASE A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(2R)-1-{[(2S,3R,5R)-1-cyclohexyl-3-hydroxy-5-{[2-(morpholin-4-yl)ethyl]carbamoyl}oct-7-yn-2-yl]amino}-3-(methylsulfa nyl)-1-oxopropan-2-yl]-1H-benzimidazole-2-carboxamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-03 | | Release date: | 2000-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
8TDU
 
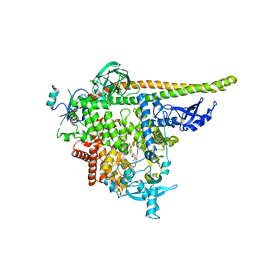 | | STX-478, a Mutant-Selective, Allosteric Inhibitor bound to PI3Kalpha | | Descriptor: | N-(2-aminopyrimidin-5-yl)-N'-[(1R)-1-(5,7-difluoro-3-methyl-1-benzofuran-2-yl)-2,2,2-trifluoroethyl]urea, N~2~-{(4S,11aP)-2-[(4S)-4-(difluoromethyl)-2-oxo-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-alaninamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Hilbert, B.J, Brooijmans, N, Buckbinder, L, St.Jean Jr, D.J. | | Deposit date: | 2023-07-05 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | STX-478, a Mutant-Selective, Allosteric PI3K alpha Inhibitor Spares Metabolic Dysfunction and Improves Therapeutic Response in PI3K alpha-Mutant Xenografts.
Cancer Discov, 13, 2023
|
|
8TGD
 
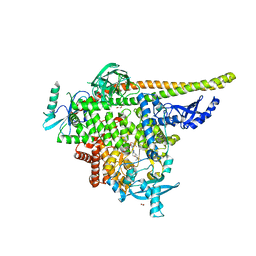 | | STX-478, a Mutant-Selective, Allosteric Inhibitor bound to H1047R PI3Kalpha | | Descriptor: | 1,2-ETHANEDIOL, N-(2-aminopyrimidin-5-yl)-N'-[(1R)-1-(5,7-difluoro-3-methyl-1-benzofuran-2-yl)-2,2,2-trifluoroethyl]urea, N~2~-{(4S,11aP)-2-[(4S)-4-(difluoromethyl)-2-oxo-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-alaninamide, ... | | Authors: | Hilbert, B, Brooijmans, N, Buckbinder, L, St.Jean Jr, D.J. | | Deposit date: | 2023-07-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.928 Å) | | Cite: | STX-478, a Mutant-Selective, Allosteric PI3K alpha Inhibitor Spares Metabolic Dysfunction and Improves Therapeutic Response in PI3K alpha-Mutant Xenografts.
Cancer Discov, 13, 2023
|
|
3OXE
 
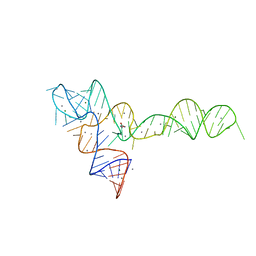 | | crystal structure of glycine riboswitch, Mn2+ soaked | | Descriptor: | GLYCINE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OWZ
 
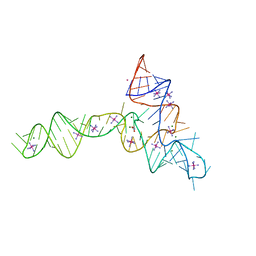 | | Crystal structure of glycine riboswitch, soaked in Iridium | | Descriptor: | Domain II of glycine riboswitch, GLYCINE, IRIDIUM HEXAMMINE ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-20 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.949 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OXB
 
 | |
3OXM
 
 | | crystal structure of glycine riboswitch, Tl-Acetate soaked | | Descriptor: | GLYCINE, MAGNESIUM ION, THALLIUM (I) ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OXJ
 
 | | crystal structure of glycine riboswitch, soaked in Ba2+ | | Descriptor: | BARIUM ION, GLYCINE, MAGNESIUM ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3P04
 
 | | Crystal Structure of the BCR protein from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR8 | | Descriptor: | Uncharacterized BCR | | Authors: | Vorobiev, S, Lew, S, Seetharaman, J, Hamilton, H, Xiao, R, Patel, D.J, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-27 | | Release date: | 2010-10-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the BCR protein from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR8.
To be Published
|
|
3OMV
 
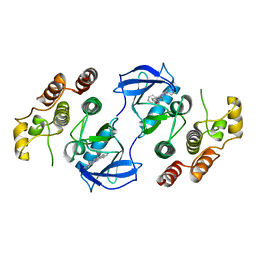 | | Crystal structure of c-raf (raf-1) | | Descriptor: | (1E)-5-(1-piperidin-4-yl-3-pyridin-4-yl-1H-pyrazol-4-yl)-2,3-dihydro-1H-inden-1-one oxime, RAF proto-oncogene serine/threonine-protein kinase | | Authors: | Hatzivassiliou, G, Song, K, Yen, I, Brandhuber, B.J, Anderson, D.J, Alvarado, R, Ludlam, M.J, Stokoe, D, Gloor, S.L, Vigers, G.P.A, Morales, T, Aliagas, I, Liu, B, Sideris, S, Hoeflich, K.P, Jaiswal, B.S, Seshagiri, S, Koeppen, H, Belvin, M, Friedman, L.S, Malek, S. | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth.
Nature, 464, 2010
|
|
3PTA
 
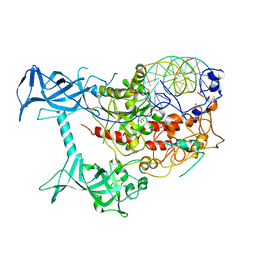 | | Crystal structure of human DNMT1(646-1600) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
3PMQ
 
 | |
3Q0B
 
 | |
3Q0F
 
 | | Crystal structure of SUVH5 SRA- methylated CHH DNA complex | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*GP*GP*AP*GP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*(5CM)P*CP*TP*CP*AP*G)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Eerappa, R, Simanshu, D.K, Patel, D.J. | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
3PT9
 
 | | Crystal structure of mouse DNMT1(731-1602) in the free state | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
3Q0D
 
 | | Crystal structure of SUVH5 SRA- hemi methylated CG DNA complex | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*TP*GP*AP*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*AP*(5CM)P*GP*TP*CP*AP*G)-3'), ... | | Authors: | Eerappa, R, Simanshu, D.K, Patel, D.J. | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3704 Å) | | Cite: | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
3QY6
 
 | | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases | | Descriptor: | FE (III) ION, MAGNESIUM ION, Tyrosine-protein phosphatase YwqE | | Authors: | Kim, H.S, Lee, S.J, Yoon, H.J, An, D.R, Kim, D.J, Kim, S.-J, Suh, S.W. | | Deposit date: | 2011-03-03 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of YwqE from Bacillus subtilis and CpsB from Streptococcus pneumoniae, unique metal-dependent tyrosine phosphatases.
J.Struct.Biol., 175, 2011
|
|
3QZT
 
 | | Crystal Structure of BPTF bromo in complex with histone H4K16ac - Form II | | Descriptor: | GLYCEROL, Histone H4, Nucleosome-remodeling factor subunit BPTF | | Authors: | Li, H, Ruthenburg, A.J, Patel, D.J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
3QZS
 
 | |
3RB3
 
 | |
3RBD
 
 | |
3QT3
 
 | | Analysis of a New Family of Widely Distributed Metal-independent alpha-Mannosidases Provides Unique Insight into the Processing of N-linked Glycans, Clostridium perfringens CPE0426 apo-structure | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein CPE0426 | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
