1FC0
 
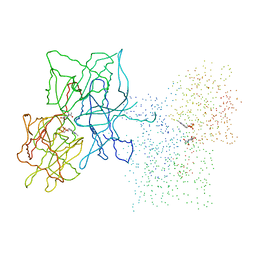 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE COMPLEXED WITH N-ACETYL-BETA-D-GLUCOPYRANOSYLAMINE | | Descriptor: | GLYCOGEN PHOSPHORYLASE, LIVER FORM, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Rath, V.L, Ammirati, M, LeMotte, P.K, Fennell, K.F, Mansour, M.M, Danley, D.E, Hynes, T.R, Schulte, G.K, Wasilko, D.J, Pandit, J. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation of human liver glycogen phosphorylase by alteration of the secondary structure and packing of the catalytic core.
Mol.Cell, 6, 2000
|
|
2M79
 
 | | [Asp2,11]RTD-1 | | Descriptor: | [Asp2,11]RTD-1 | | Authors: | Conibear, A.C, Bochen, A, Rosengren, K, Kessler, H, Craik, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2014-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Cyclic Cystine Ladder of Theta-Defensins as a Stable, Bifunctional Scaffold: A Proof-of-Concept Study Using the Integrin-Binding RGD Motif
Chembiochem, 15, 2014
|
|
1BXX
 
 | |
1BIV
 
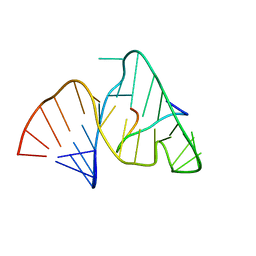 | | BOVINE IMMUNODEFICIENCY VIRUS TAT-TAR COMPLEX, NMR, 5 STRUCTURES | | Descriptor: | TAR RNA, TAT PEPTIDE | | Authors: | Ye, X, Kumar, R.A, Patel, D.J. | | Deposit date: | 1996-06-12 | | Release date: | 1996-12-23 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Molecular recognition in the bovine immunodeficiency virus Tat peptide-TAR RNA complex.
Chem.Biol., 2, 1995
|
|
1J77
 
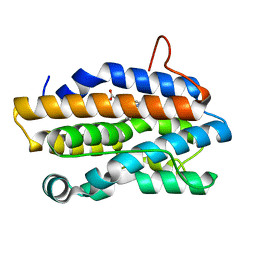 | | Crystal Structure of Gram-negative Bacterial Heme Oxygenase Complexed with Heme | | Descriptor: | HemO, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schuller, D.J, Zhu, W, Stojiljkovic, I, Wilks, A, Poulos, T.L. | | Deposit date: | 2001-05-15 | | Release date: | 2001-05-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of heme oxygenase from the gram-negative pathogen Neisseria meningitidis and a comparison with mammalian heme oxygenase-1.
Biochemistry, 40, 2001
|
|
2N1L
 
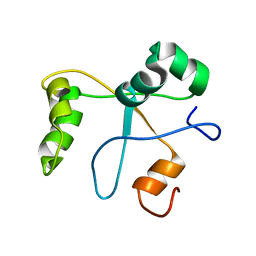 | | Solution structure of the BCOR PUFD | | Descriptor: | BCL-6 corepressor | | Authors: | Wong, S.J, Gearhart, M.D, Ha, D.J, Corcoran, C.M, Diaz, V, Taylor, A.B, Schirf, V, Ilangovan, U, Hinck, A.P, Demeler, B, Hart, J, Bardwell, V.J, Kim, C.A. | | Deposit date: | 2015-04-06 | | Release date: | 2016-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the hierarchical assembly of the core of PRC1.1
To be Published
|
|
1C0Y
 
 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT POSITIONED OPPOSITE DA AT A TEMPLATE-PRIMER JUNCTION | | Descriptor: | 2-AMINOFLUORENE, DNA (5'-D(*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*GP*TP*AP*GP*C)-3') | | Authors: | Gu, Z, Gorin, A, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1999-07-19 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of aminofluorene [AF]-stacked conformers of the syn [AF]-C8-dG adduct positioned opposite dC or dA at a template-primer junction.
Biochemistry, 38, 1999
|
|
1IFT
 
 | | RICIN A-CHAIN (RECOMBINANT) | | Descriptor: | RICIN | | Authors: | Weston, S.A, Tucker, A.D, Thatcher, D.R, Derbyshire, D.J, Pauptit, R.A. | | Deposit date: | 1996-07-05 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of recombinant ricin A-chain at 1.8 A resolution.
J.Mol.Biol., 244, 1994
|
|
1BN0
 
 | | SL3 HAIRPIN FROM THE PACKAGING SIGNAL OF HIV-1, NMR, 11 STRUCTURES | | Descriptor: | SL3 RNA HAIRPIN | | Authors: | Pappalardo, L, Kerwood, D.J, Pelczer, I, Borer, P.N. | | Deposit date: | 1998-07-31 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional folding of an RNA hairpin required for packaging HIV-1.
J.Mol.Biol., 282, 1998
|
|
2N0N
 
 | | NMR solution structure for lactam (5,9) 11mer | | Descriptor: | lactam (5,9) 11mer peptide | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-10 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
7JNN
 
 | |
7JN6
 
 | |
2J9W
 
 | | Structural insight into the ESCRT-I-II link and its role in MVB trafficking | | Descriptor: | VPS28-PROV PROTEIN | | Authors: | Gill, D.J, Teo, H.L, Sun, J, Perisic, O, Veprintsev, D.B, Emr, S.D, Williams, R.L. | | Deposit date: | 2006-11-16 | | Release date: | 2007-01-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Insight Into the Escrt-I/-II Link and its Role in Mvb Trafficking.
Embo J., 26, 2007
|
|
7KTQ
 
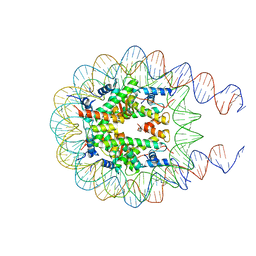 | | Nucleosome from a dimeric PRC2 bound to a nucleosome | | Descriptor: | 601 DNA (167-MER), Histone H2A, Histone H2B, ... | | Authors: | Grau, D.J, Armache, K.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
2J9U
 
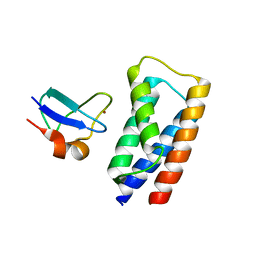 | | 2 Angstrom X-ray structure of the yeast ESCRT-I Vps28 C-terminus in complex with the NZF-N domain from ESCRT-II | | Descriptor: | VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 28, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 36, ZINC ION | | Authors: | Gill, D.J, Teo, H.L, Sun, J, Perisic, O, Veprintsev, D.B, Emr, S.D, Williams, R.L. | | Deposit date: | 2006-11-16 | | Release date: | 2007-01-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight Into the Escrt-I/-II Link and its Role in Mvb Trafficking.
Embo J., 26, 2007
|
|
7JW3
 
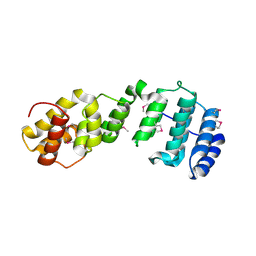 | | Crystal structure of Aedes aegypti Nibbler NTD domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1C01
 
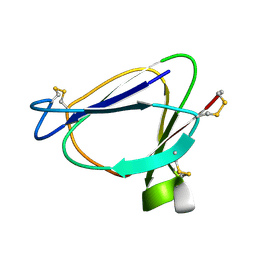 | | SOLUTION STRUCTURE OF MIAMP1, A PLANT ANTIMICROBIAL PROTEIN | | Descriptor: | ANTIMICROBIAL PEPTIDE 1 | | Authors: | McManus, A.M, Nielsen, K.J, Marcus, J.P, Harrison, S.J, Green, J.L, Manners, J.M, Craik, D.J. | | Deposit date: | 1999-07-13 | | Release date: | 2000-07-19 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | MiAMP1, a novel protein from Macadamia integrifolia adopts a Greek key beta-barrel fold unique amongst plant antimicrobial proteins.
J.Mol.Biol., 293, 1999
|
|
1DT7
 
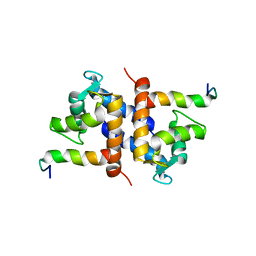 | |
2K77
 
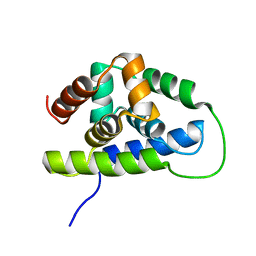 | | NMR solution structure of the Bacillus subtilis ClpC N-domain | | Descriptor: | Negative regulator of genetic competence clpC/mecB | | Authors: | Kojetin, D.J, McLaughlin, P.D, Thompson, R.J, Rance, M, Cavanagh, J. | | Deposit date: | 2008-08-04 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and motional contributions of the Bacillus subtilis ClpC N-domain to adaptor protein interactions.
J.Mol.Biol., 387, 2009
|
|
7KBG
 
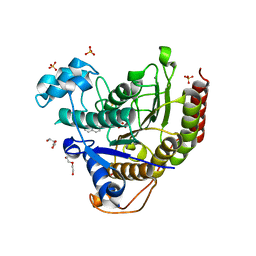 | |
1G8X
 
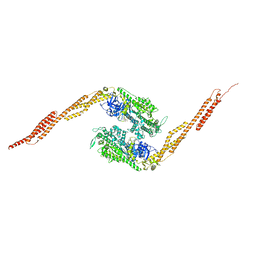 | | STRUCTURE OF A GENETICALLY ENGINEERED MOLECULAR MOTOR | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN II HEAVY CHAIN FUSED TO ALPHA-ACTININ 3 | | Authors: | Kliche, W, Fujita-Becker, S, Kollmar, M, Manstein, D.J, Kull, F.J. | | Deposit date: | 2000-11-21 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a genetically engineered molecular motor.
EMBO J., 20, 2001
|
|
2KF4
 
 | | Barnase high pressure structure | | Descriptor: | Ribonuclease | | Authors: | Williamson, M.P, Wilton, D.J. | | Deposit date: | 2009-02-11 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Pressure-dependent structure changes in barnase on ligand binding reveal intermediate rate fluctuations.
Biophys.J., 97, 2009
|
|
2KF6
 
 | |
1DML
 
 | | CRYSTAL STRUCTURE OF HERPES SIMPLEX UL42 BOUND TO THE C-TERMINUS OF HSV POL | | Descriptor: | DNA POLYMERASE, DNA POLYMERASE PROCESSIVITY FACTOR | | Authors: | Zuccola, H.J, Filman, D.J, Coen, D.M, Hogle, J.M. | | Deposit date: | 1999-12-14 | | Release date: | 2000-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase.
Mol.Cell, 5, 2000
|
|
7K24
 
 | |
