1DJE
 
 | | CRYSTAL STRUCTURE OF THE PLP-BOUND FORM OF 8-AMINO-7-OXONANOATE SYNTHASE | | Descriptor: | 8-AMINO-7-OXONANOATE SYNTHASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Webster, S.P, Alexeev, D, Campopiano, D.J, Watt, R.M, Alexeeva, M, Sawyer, L, Baxter, R.L. | | Deposit date: | 1999-12-02 | | Release date: | 2000-12-04 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Mechanism of 8-amino-7-oxononanoate synthase: spectroscopic, kinetic, and crystallographic studies.
Biochemistry, 39, 2000
|
|
1DD6
 
 | | IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH A MERCAPTOCARBOXYLATE INHIBITOR | | Descriptor: | (2-MERCAPTOMETHYL-4-PHENYL-BUTYRYLIMINO)-(5-TETRAZOL-1-YLMETHYL-THIOPHEN-2-YL)-ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, SULFATE ION, ... | | Authors: | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
1DJS
 
 | |
1DVN
 
 | |
1DF6
 
 | |
1DJ9
 
 | | CRYSTAL STRUCTURE OF 8-AMINO-7-OXONANOATE SYNTHASE (OR 7-KETO-8AMINIPELARGONATE OR KAPA SYNTHASE) COMPLEXED WITH PLP AND THE PRODUCT 8(S)-AMINO-7-OXONANONOATE (OR KAPA). THE ENZYME OF BIOTIN BIOSYNTHETIC PATHWAY. | | Descriptor: | 8-AMINO-7-OXONONANOATE SYNTHASE, MAGNESIUM ION, N-[7-KETO-8-AMINOPELARGONIC ACID]-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Webster, S.P, Alexeev, D, Campopiano, D.J, Watt, R.M, Alexeeva, M, Sawyer, L, Baxter, R.L. | | Deposit date: | 1999-12-02 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of 8-amino-7-oxononanoate synthase: spectroscopic, kinetic, and crystallographic studies.
Biochemistry, 39, 2000
|
|
1DT7
 
 | |
1DML
 
 | | CRYSTAL STRUCTURE OF HERPES SIMPLEX UL42 BOUND TO THE C-TERMINUS OF HSV POL | | Descriptor: | DNA POLYMERASE, DNA POLYMERASE PROCESSIVITY FACTOR | | Authors: | Zuccola, H.J, Filman, D.J, Coen, D.M, Hogle, J.M. | | Deposit date: | 1999-12-14 | | Release date: | 2000-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase.
Mol.Cell, 5, 2000
|
|
1E42
 
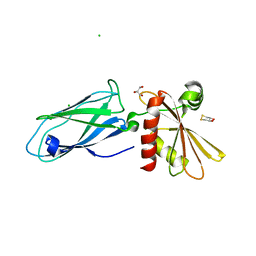 | | Beta2-adaptin appendage domain, from clathrin adaptor AP2 | | Descriptor: | AP-2 COMPLEX SUBUNIT BETA, CHLORIDE ION, DITHIANE DIOL, ... | | Authors: | Owen, D.J, Evans, P.R, McMahon, H.T. | | Deposit date: | 2000-06-27 | | Release date: | 2000-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure and Function of the Beta2-Adaptin Appendage Domain
Embo J., 19, 2000
|
|
1DSV
 
 | | STRUCTURE OF THE MMTV NUCLEOCAPSID PROTEIN (C-TERMINAL ZINC FINGER) | | Descriptor: | NUCLEIC ACID BINDING PROTEIN P14, ZINC ION | | Authors: | Klein, D.J, Johnson, P.E, Zollars, E.S, De Guzman, R.N, Summers, M.F. | | Deposit date: | 2000-01-08 | | Release date: | 2000-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle.
Biochemistry, 39, 2000
|
|
1DSQ
 
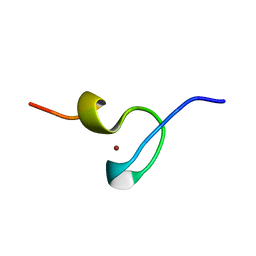 | | STRUCTURE OF THE MMTV NUCLEOCAPSID PROTEIN (ZINC FINGER 1) | | Descriptor: | NUCLEIC ACID BINDING PROTEIN P14, ZINC ION | | Authors: | Klein, D.J, Johnson, P.E, Zollars, E.S, De Guzman, R.N, Summers, M.F. | | Deposit date: | 2000-01-08 | | Release date: | 2000-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle.
Biochemistry, 39, 2000
|
|
1DTO
 
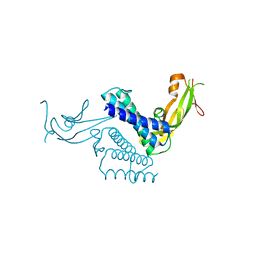 | | CRYSTAL STRUCTURE OF THE COMPLETE TRANSACTIVATION DOMAIN OF E2 PROTEIN FROM THE HUMAN PAPILLOMAVIRUS TYPE 16 | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Antson, A.A, Burns, J.E, Moroz, O.V, Scott, D.J, Sanders, C.M, Bronstein, I.B, Dodson, G.G, Wilson, K.S, Maitland, N. | | Deposit date: | 2000-01-13 | | Release date: | 2000-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the intact transactivation domain of the human papillomavirus E2 protein.
Nature, 403, 2000
|
|
1DVM
 
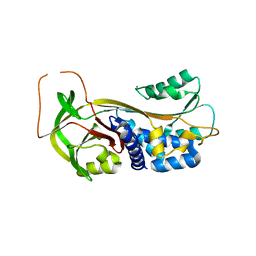 | | ACTIVE FORM OF HUMAN PAI-1 | | Descriptor: | CHLORIDE ION, PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Stout, T.J, Graham, H, Buckley, D.I, Matthews, D.J. | | Deposit date: | 2000-01-21 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of active and latent PAI-1: a possible stabilizing role for chloride ions.
Biochemistry, 39, 2000
|
|
1DPS
 
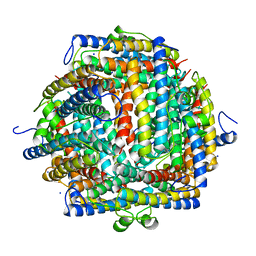 | | THE CRYSTAL STRUCTURE OF DPS, A FERRITIN HOMOLOG THAT BINDS AND PROTECTS DNA | | Descriptor: | DPS, SODIUM ION | | Authors: | Grant, R.A, Filman, D.J, Finkel, S.E, Kolter, R, Hogle, J.M. | | Deposit date: | 1998-02-23 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Dps, a ferritin homolog that binds and protects DNA.
Nat.Struct.Biol., 5, 1998
|
|
1EVH
 
 | |
1EBV
 
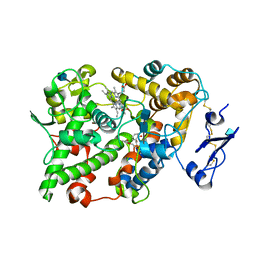 | | OVINE PGHS-1 COMPLEXED WITH SALICYL HYDROXAMIC ACID | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID SALICYLOYL-AMINO-ESTER, ... | | Authors: | Loll, P.J, Sharkey, C.T, O'Connor, S.J, Fitzgerald, D.J. | | Deposit date: | 2000-01-24 | | Release date: | 2000-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | O-acetylsalicylhydroxamic acid, a novel acetylating inhibitor of prostaglandin H2 synthase: structural and functional characterization of enzyme-inhibitor interactions.
Mol.Pharmacol., 60, 2001
|
|
1EJI
 
 | | RECOMBINANT SERINE HYDROXYMETHYLTRANSFERASE (MOUSE) | | Descriptor: | 5-HYDROXYMETHYLENE-6-HYDROFOLIC ACID, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], SERINE HYDROXYMETHYLTRANSFERASE | | Authors: | Szebenyi, D.M.E, Liu, X, Kriksunov, I.A, Stover, P.J, Thiel, D.J. | | Deposit date: | 2000-03-02 | | Release date: | 2000-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a murine cytoplasmic serine hydroxymethyltransferase quinonoid ternary complex: evidence for asymmetric obligate dimers.
Biochemistry, 39, 2000
|
|
1E6I
 
 | |
1EEG
 
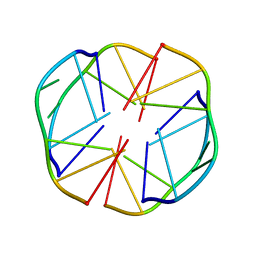 | | A(GGGG)A HEXAD PAIRING ALIGMENT FOR THE D(G-G-A-G-G-A-G) SEQUENCE | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*A)-3') | | Authors: | Kettani, A, Gorin, A, Majumdar, A, Hermann, T, Skripkin, E, Zhao, H, Jones, R, Patel, D.J. | | Deposit date: | 2000-01-31 | | Release date: | 2000-04-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A dimeric DNA interface stabilized by stacked A.(G.G.G.G).A hexads and coordinated monovalent cations.
J.Mol.Biol., 297, 2000
|
|
1EM6
 
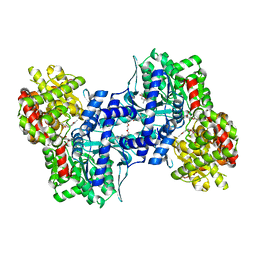 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE A COMPLEXED WITH GLCNAC AND CP-526,423 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BIS[5-CHLORO-1H-INDOL-2-YL-CARBONYL-AMINOETHYL]-ETHYLENE GLYCOL, LIVER GLYCOGEN PHOSPHORYLASE, ... | | Authors: | Rath, V.L, Ammirati, M, Danley, D.E, Ekstrom, J.L, Hynes, T.R, Olson, T.V, Hoover, D.J. | | Deposit date: | 2000-03-16 | | Release date: | 2000-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human liver glycogen phosphorylase inhibitors bind at a new allosteric site.
Chem.Biol., 7, 2000
|
|
1EXV
 
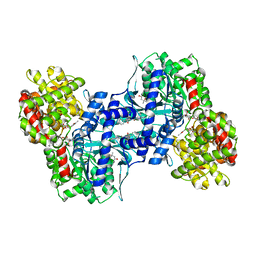 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE A COMPLEXED WITH GLCNAC AND CP-403,700 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, LIVER GLYCOGEN PHOSPHORYLASE, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Rath, V.L, Ammirati, M, Danley, D.E, Ekstrom, J.L, Hynes, T.R, Olson, T.V, Hoover, D.J. | | Deposit date: | 2000-05-04 | | Release date: | 2000-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human liver glycogen phosphorylase inhibitors bind at a new allosteric site.
Chem.Biol., 7, 2000
|
|
1EXY
 
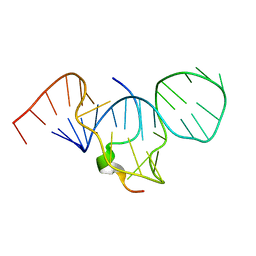 | | SOLUTION STRUCTURE OF HTLV-1 PEPTIDE BOUND TO ITS RNA APTAMER TARGET | | Descriptor: | HTLV-1 REX PEPTIDE, RNA APTAMER, 33-MER | | Authors: | Jiang, F, Gorin, A, Hu, W, Majumdar, A, Baskerville, S, Xu, W, Ellington, A, Patel, D.J. | | Deposit date: | 2000-05-05 | | Release date: | 2000-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Anchoring an extended HTLV-1 Rex peptide within an RNA major groove containing junctional base triples.
Structure Fold.Des., 7, 1999
|
|
1F40
 
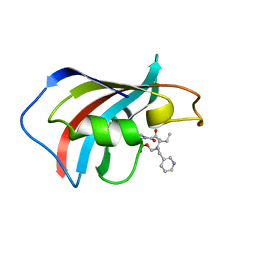 | | SOLUTION STRUCTURE OF FKBP12 COMPLEXED WITH GPI-1046, A NEUROTROPHIC LIGAND | | Descriptor: | (2S)-[3-PYRIDYL-1-PROPYL]-1-[3,3-DIMETHYL-1,2-DIOXOPENTYL]-2-PYRROLIDINECARBOXYLATE, FK506 BINDING PROTEIN (FKBP12) | | Authors: | Sich, C, Improta, S, Cowley, D.J, Guenet, C, Merly, J.P, Teufel, M, Saudek, V. | | Deposit date: | 2000-06-07 | | Release date: | 2000-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a neurotrophic ligand bound to FKBP12 and its effects on protein dynamics.
Eur.J.Biochem., 267, 2000
|
|
1F4U
 
 | | THERMOPHILIC P450: CYP119 FROM SULFOLOBUS SOLFACTARICUS | | Descriptor: | CYTOCHROME P450 119, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yano, J.K, Koo, L.S, Schuller, D.J, Li, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2000-06-09 | | Release date: | 2000-10-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of a thermophilic cytochrome P450 from the archaeon Sulfolobus solfataricus.
J.Biol.Chem., 275, 2000
|
|
1F3S
 
 | | Solution Structure of DNA Sequence GGGTTCAGG Forms GGGG Tetrade and G(C-A) Triad. | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*TP*CP*AP*GP*G)-3') | | Authors: | Kettani, A, Basu, G, Gorin, A, Majumdar, A, Skripkin, E, Patel, D.J. | | Deposit date: | 2000-06-06 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A two-stranded template-based approach to G.(C-A) triad formation: designing novel structural elements into an existing DNA framework.
J.Mol.Biol., 301, 2000
|
|
