6OJU
 
 | | Crystal structure of human thymidylate synthase Delta (7-29) in complex with dUMP and 2-amino-4-oxo-4,7-dihydro-pyrrolo[2,3-d]pyrimidine-methyl-phenyl-D-glutamic acid | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, N-{4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}-D-glutamic acid, Thymidylate synthase,Thymidylate synthase | | Authors: | Czyzyk, D.J, Anderson, K.S, Valhondo, M, Jorgensen, W.L. | | Deposit date: | 2019-04-12 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.884 Å) | | Cite: | Understanding the structural basis of species selective, stereospecific inhibition for Cryptosporidium and human thymidylate synthase.
Febs Lett., 593, 2019
|
|
4AZ7
 
 | | Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, BETA-N-ACETYLHEXOSAMINIDASE, ... | | Authors: | Pluvinage, B, Stubbs, K.A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-06-23 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of the Family 20 Glycoside Hydrolase Catalytic Modules in the Streptococcus Pneumoniae Exo-Beta-D-N-Acetylglucosaminidase, Strh.
Org.Biomol.Chem., 11, 2013
|
|
4AZH
 
 | | Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH | | Descriptor: | 1,2-ETHANEDIOL, BETA-N-ACETYLHEXOSAMINIDASE, N-acetylglucosaminono-1,5-lactone (Z)-oxime | | Authors: | Pluvinage, B, Stubbs, K.A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-06-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Inhibition of the Family 20 Glycoside Hydrolase Catalytic Modules in the Streptococcus Pneumoniae Exo-Beta-D-N-Acetylglucosaminidase, Strh.
Org.Biomol.Chem., 11, 2013
|
|
6USX
 
 | | Identification of the Clinical Development Candidate MRTX849, a Covalent KRASG12C Inhibitor for the Treatment of Cancer | | Descriptor: | 1-{4-[2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}-7-(naphthalen-1-yl)-5,6,7,8-tetrahydropyrido[3,4-d]pyrimidin-4-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vigers, G.P, Smith, D.J. | | Deposit date: | 2019-10-28 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Identification of the Clinical Development CandidateMRTX849, a Covalent KRASG12CInhibitor for the Treatment of Cancer.
J.Med.Chem., 63, 2020
|
|
6OPF
 
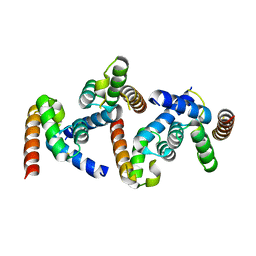 | |
3K54
 
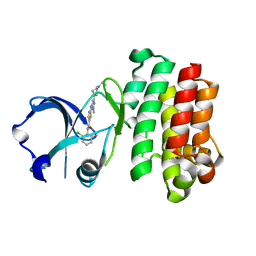 | | Structures of human Bruton's tyrosine kinase in active and inactive conformations suggests a mechanism of activation for TEC family kinases. | | Descriptor: | N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, Tyrosine-protein kinase BTK | | Authors: | Marcotte, D.J, Liu, Y.-T, Arduini, R.M, Hession, C.A, Miatkowski, K, Wildes, C.P, Cullen, P.F, Hopkins, B.T, Mertsching, E, Jenkins, T.J, Romanowski, M.J, Baker, D.P, Silvian, L.F. | | Deposit date: | 2009-10-06 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of human Bruton's tyrosine kinase in active and inactive conformations suggest a mechanism of activation for TEC family kinases.
Protein Sci., 19, 2010
|
|
1L5S
 
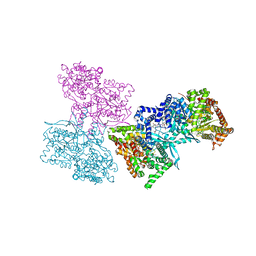 | | Human liver glycogen phosphorylase complexed with uric acid, N-Acetyl-beta-D-glucopyranosylamine, and CP-403,700 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Glycogen phosphorylase, liver form, ... | | Authors: | Ekstrom, J.L, Pauly, T.A, Carty, M.D, Soeller, W.C, Culp, J, Danley, D.E, Hoover, D.J, Treadway, J.L, Gibbs, E.M, Fletterick, R.J, Day, Y.S.N, Myszka, D.G, Rath, V.L. | | Deposit date: | 2002-03-07 | | Release date: | 2002-12-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity analysis of the purine binding
site of human liver glycogen phosphorylase.
Chem.Biol., 9, 2002
|
|
6O6X
 
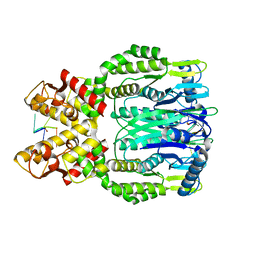 | |
4AZG
 
 | | Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BETA-N-ACETYLHEXOSAMINIDASE, ... | | Authors: | Pluvinage, B, Stubbs, K.A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-06-25 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of the Family 20 Glycoside Hydrolase Catalytic Modules in the Streptococcus Pneumoniae Exo-Beta-D-N-Acetylglucosaminidase, Strh.
Org.Biomol.Chem., 11, 2013
|
|
7SDE
 
 | | Cryo-EM structure of Nse5/6 heterodimer | | Descriptor: | Non-structural maintenance of chromosome element 5, Ubiquitin-like protein SMT3,DNA repair protein KRE29 chimera | | Authors: | Yu, Y, Patel, D.J, Zhao, X.L. | | Deposit date: | 2021-09-29 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The cryo-EM structure of Nse5/6 complex with the C terminal part of Nse5
To Be Published
|
|
4CFU
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 3-[2-azanyl-6-(cyclohexylmethoxy)-7H-purin-8-yl]-2-methyl-benzoic acid, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2, ... | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4CFM
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 6-(cyclohexylmethoxy)-8-(2-methylphenyl)-9H-purin-2-amine, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-18 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
3J9F
 
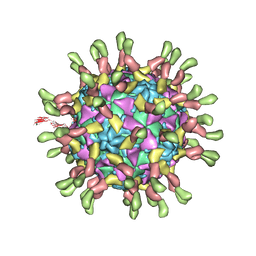 | | Poliovirus complexed with soluble, deglycosylated poliovirus receptor (Pvr) at 4 degrees C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-02-11 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
4CC6
 
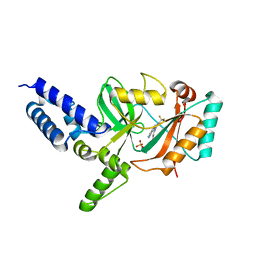 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-{[2-(1H-pyrazolo[3,4-c]pyridin-3-yl)-6-(trifluoromethyl)pyridin-4-yl]amino}ethanol, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
6PC5
 
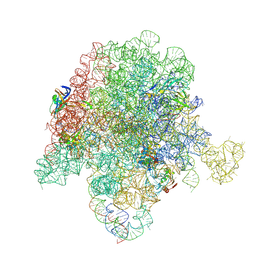 | | E. coli 50S ribosome bound to compounds 46 and VS1 | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-4,12-dimethyl-1,7,22-trioxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl isoquinolin-3-ylcarbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-16 | | Release date: | 2020-06-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
5T1Y
 
 | | MLA10 coiled-coil fragment | | Descriptor: | MLA10, TRIETHYLENE GLYCOL | | Authors: | Williams, S.J, Kobe, B, Bentham, A, Ericsson, D.J. | | Deposit date: | 2016-08-22 | | Release date: | 2016-11-02 | | Last modified: | 2016-11-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The CC domain structure from the wheat stem rust resistance protein Sr33 challenges paradigms for dimerization in plant NLR proteins.
Proc.Natl.Acad.Sci.USA, 2016
|
|
6PF7
 
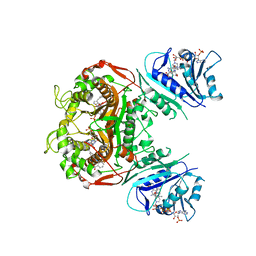 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP and 2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)benzoic acid | | Descriptor: | 2-({4-[(2-amino-4-oxo-4,7-dihydro-1H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)benzoic acid, 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Czyzyk, D.J, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
3K8D
 
 | | Crystal structure of E. coli lipopolysaccharide specific CMP-KDO synthetase in complex with CTP and 2-deoxy-Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, 3-deoxy-manno-octulosonate cytidylyltransferase, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Heyes, D.J, Levy, C.W, Lafite, P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based mechanism of CMP-2-keto-3-deoxymanno-octulonic acid synthetase: convergent evolution of a sugar-activating enzyme with DNA/RNA polymerases
J.Biol.Chem., 284, 2009
|
|
4C2Z
 
 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA and inhibitor bound | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Thinon, E, Serwa, R.A, Brannigan, J.A, Brassat, U, Wright, M.H, Heal, W.P, Wilkinson, A.J, Mann, D.J, Tate, E.W. | | Deposit date: | 2013-08-20 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Chemical Proteomics Defines the Mammalian N- Myristoylated Proteome in Live Cells Global Profiling of Co- and Post-Translationally N-Myristoylated Proteomes in Human Cells.
Nat.Commun., 5, 2014
|
|
7R76
 
 | | Cryo-EM structure of DNMT5 in apo state | | Descriptor: | DNA repair protein Rad8, ZINC ION | | Authors: | Wang, J, Patel, D.J. | | Deposit date: | 2021-06-24 | | Release date: | 2022-02-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into DNMT5-mediated ATP-dependent high-fidelity epigenome maintenance.
Mol.Cell, 82, 2022
|
|
7R77
 
 | |
7R78
 
 | |
4PDZ
 
 | | Crystal Structure of Calcium-loaded S100B bound to SBi4172 | | Descriptor: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Pierce, A.D, Wilder, P.T, Neau, D, Toth, E.A, Weber, D.J. | | Deposit date: | 2014-04-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Covalent Small Molecule Inhibitors of Ca(2+)-Bound S100B.
Biochemistry, 53, 2014
|
|
4PE4
 
 | | Crystal Structure of Calcium-loaded S100B bound to SC1475 | | Descriptor: | 2,3-dimethoxy-5-[(1S)-1-phenylpropyl]benzene-1,4-diol, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Pierce, A.D, Wilder, P.T, Neau, D, Toth, E.A, Weber, D.J. | | Deposit date: | 2014-04-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.178 Å) | | Cite: | Covalent Small Molecule Inhibitors of Ca(2+)-Bound S100B.
Biochemistry, 53, 2014
|
|
6PCS
 
 | | E. coli 50S ribosome bound to compound 40e | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,26aR)-14-hydroxy-4,12-dimethyl-1,7,16,22-tetraoxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl [4-(trifluoromethyl)phenyl]carbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-06-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
