6ZJO
 
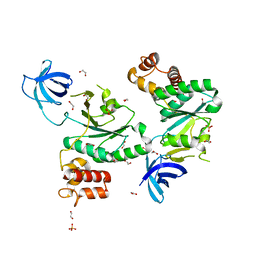 | | Crystal Structure of Staphylococcus aureus RsgA. | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Small ribosomal subunit biogenesis GTPase RsgA, ... | | Authors: | Bennison, D.J, Rafferty, J.B, Corrigan, R.M. | | Deposit date: | 2020-06-29 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Stringent Response Inhibits 70S Ribosome Formation in Staphylococcus aureus by Impeding GTPase-Ribosome Interactions.
Mbio, 12, 2021
|
|
6ZHL
 
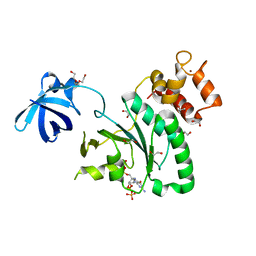 | | Crystal Structure of Staphylococcus aureus RsgA bound to ppGpp. | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5',3'-TETRAPHOSPHATE, Small ribosomal subunit biogenesis GTPase RsgA, ... | | Authors: | Bennison, D.J, Rafferty, J.B, Corrigan, R.M. | | Deposit date: | 2020-06-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Stringent Response Inhibits 70S Ribosome Formation in Staphylococcus aureus by Impeding GTPase-Ribosome Interactions.
Mbio, 12, 2021
|
|
4MHW
 
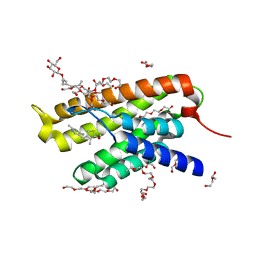 | | Crystal structure of ThiT with small molecule BAT-25 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 2-{4-[(4-amino-2-methylpyrimidin-5-yl)methyl]-3-methylthiophen-2-yl}ethanol, ... | | Authors: | Swier, L.J.Y.M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2013-08-30 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of ThiT with small molecules modulators
To be Published
|
|
7A96
 
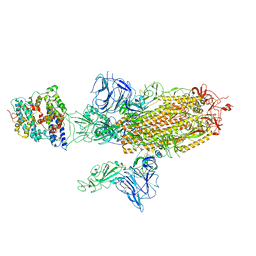 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound and 1 RBD Erect in Anticlockwise Direction | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A97
 
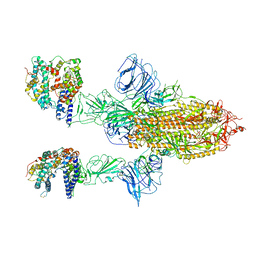 | | SARS-CoV-2 Spike Glycoprotein with 2 ACE2 Bound | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A93
 
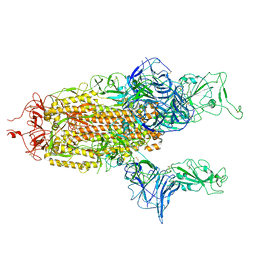 | |
4NEC
 
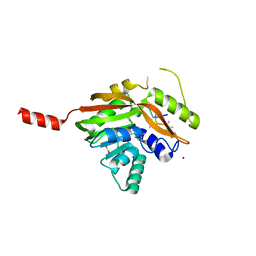 | | Conversion of a Disulfide Bond into a Thioacetal Group during Echinomycin Biosynthesis | | Descriptor: | 2-CARBOXYQUINOXALINE, ACETATE ION, Echinomycin, ... | | Authors: | Hotta, K, Keegan, R.M, Ranganathan, S, Fang, M, Bibby, J, Winn, M.D, Sato, M, Lian, M, Watanabe, K, Rigden, D.J, Kim, C.-Y. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conversion of a disulfide bond into a thioacetal group during echinomycin biosynthesis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NGF
 
 | |
4NGB
 
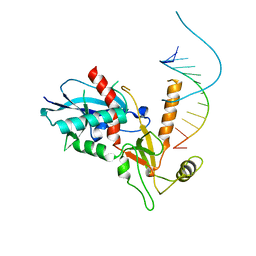 | |
4NYN
 
 | |
4NH3
 
 | |
1OYH
 
 | | Crystal Structure of P13 Alanine Variant of Antithrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin-III, ... | | Authors: | Johnson, D.J.D, Huntington, J.A. | | Deposit date: | 2003-04-04 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The influence of hinge region residue Glu-381 on antithrombin allostery and metastability
J.Biol.Chem., 279, 2004
|
|
5CNQ
 
 | | Crystal structure of the Holliday junction-resolving enzyme GEN1 (WT) in complex with product DNA, Mg2+ and Mn2+ ions | | Descriptor: | DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MANGANESE (II) ION, Nuclease-like protein, ... | | Authors: | Liu, Y.J, Freeman, A.D.J, Declais, A.C, Wilson, T.J, Gartner, A, Lilley, D.M.J. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Crystal Structure of a Eukaryotic GEN1 Resolving Enzyme Bound to DNA.
Cell Rep, 13, 2015
|
|
4NT2
 
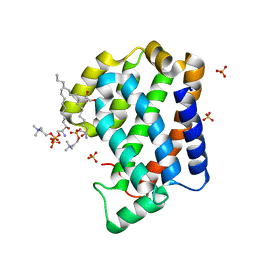 | | Crystal structure of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with lyso-sphingomyelin (d18:1) at 2.4 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(R)-{[(2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl]oxy}(hydroxy)phosphoryl]oxy}-N,N,N-trimethylethanaminium, SULFATE ION, ... | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
5CO8
 
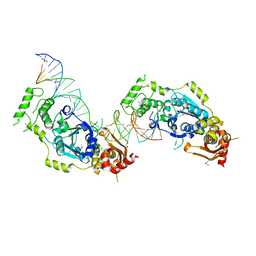 | | Crystal structure of the Holliday junction-resolving enzyme GEN1 (WT) in complex with product DNA and Mg2+ ion | | Descriptor: | DNA (31-MER), DNA (5'-D(*AP*GP*AP*CP*TP*GP*CP*AP*GP*TP*TP*GP*AP*GP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*A)-3'), ... | | Authors: | Liu, Y.J, Freeman, A.D.J, Declais, A.C, Wilson, T.J, Gartner, A, Lilley, D.M.J. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a Eukaryotic GEN1 Resolving Enzyme Bound to DNA.
Cell Rep, 13, 2015
|
|
4NTG
 
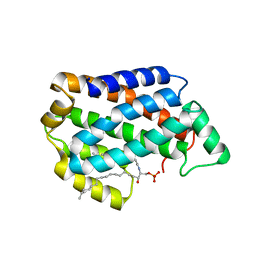 | | Crystal structure of D60A mutant of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with C12 ceramide-1-phosphate (d18:1/12:0) at 2.55 Angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(dodecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, accelerated-cell-death 11 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-12-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5505 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
4OUK
 
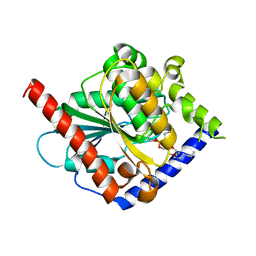 | | Crystal structure of a C6-C4 SN3 inhibited ESTERASE B from LACTOBACILLUS RHAMNOSIS | | Descriptor: | (2R)-2,3-bis(hexyloxy)propyl hydrogen (S)-pentylphosphonate, Esterase B | | Authors: | Colbert, D.A, Bennett, M.D, Lun, D.J, Holland, R, Delabre, M.-L, Loo, T.S, Anderson, B.F, Norris, G.E. | | Deposit date: | 2014-02-17 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a C6-C4 SN3 inhibited ESTERASE B from LACTOBACILLUS RHAMNOSIS
TO BE PUBLISHED
|
|
5K31
 
 | | Crystal structure of Human fibrillar procollagen type I C-propeptide Homo-trimer | | Descriptor: | CALCIUM ION, CHLORIDE ION, Collagen alpha-1(I) chain, ... | | Authors: | Sharma, U, Hulmes, D.J.S, Aghajari, N. | | Deposit date: | 2016-05-19 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of homo- and heterotrimerization of collagen I.
Nat Commun, 8, 2017
|
|
4ONQ
 
 | |
4OUD
 
 | | Engineered tyrosyl-tRNA synthetase with the nonstandard amino acid L-4,4-biphenylalanine | | Descriptor: | TYROSINE, Tyrosyl-tRNA synthetase | | Authors: | Takeuchi, R, Mandell, D.J, Lajoie, M.J, Church, G.M, Stoddard, B.L. | | Deposit date: | 2014-02-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biocontainment of genetically modified organisms by synthetic protein design.
Nature, 518, 2015
|
|
4NMH
 
 | | 11-beta-HSD1 in complex with a 3,3-Di-methyl-azetidin-2-one | | Descriptor: | (4S)-4-(2-methoxyphenyl)-3,3-dimethyl-1-[3-(methylsulfonyl)phenyl]azetidin-2-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McCoull, W, Augustin, M, Blake, C, Ertan, A, Kilgour, E.K, Krapp, S, Moore, J.E, Newcombe, N.J, Packer, M.J, Rees, A, Revill, J, Scott, J.S, Selmi, N, Gerhardt, S, Ogg, D.J, Steinbacher, S, Whittamore, P.R.O. | | Deposit date: | 2013-11-15 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification and optimisation of 3,3-dimethyl-azetidin-2-ones as potent and selective inhibitors of 11 beta-hydroxysteroid dehydrogenase type 1 (11-beta-HSD1)
TO BE PUBLISHED
|
|
7AM3
 
 | | Crystal structure of Peptiligase mutant - M222P | | Descriptor: | GLYCEROL, SULFATE ION, Subtilisin BPN' | | Authors: | Rozeboom, H.J, Janssen, D.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | From thiol-subtilisin to omniligase: Design and structure of a broadly applicable peptide ligase.
Comput Struct Biotechnol J, 19, 2021
|
|
7AM5
 
 | |
7AM6
 
 | |
7AM8
 
 | | Crystal structure of Omniligase mutant W189F | | Descriptor: | ACRYLIC ACID, CHLORIDE ION, HISTIDINE, ... | | Authors: | Rozeboom, H.J, Janssen, D.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | From thiol-subtilisin to omniligase: Design and structure of a broadly applicable peptide ligase.
Comput Struct Biotechnol J, 19, 2021
|
|
