5EZR
 
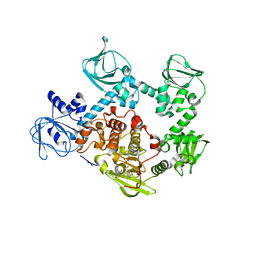 | | Crystal Structure of PVX_084705 bound to compound | | Descriptor: | CHLORIDE ION, N-[5-(3-{2-[(cyclopropylmethyl)amino]pyrimidin-4-yl}-7-[(dimethylamino)methyl]-6-methylimidazo[1,2-a]pyridin-2-yl)-2-fluorophenyl]methanesulfonamide, cGMP-dependent protein kinase, ... | | Authors: | El Bakkouri, M, Amani, M, Walker, J.R, Osborne, S, Large, J.M, Birchall, K, Bouloc, N, Smiljanic-Hurley, E, Wheldon, M, Harding, D.J, Merritt, A.T, Ansell, K.H, Coombs, P.J, Kettleborough, C.A, Stewart, B.L, Bowyer, P.W, Gutteridge, W.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Baker, D.A, Hui, R, Loppnau, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-26 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of PVX_084705 bound to compound
To Be Published
|
|
2WMJ
 
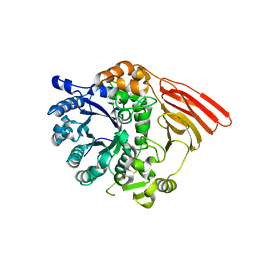 | | Crystal structure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 (Sp3GH98) in complex with the B-trisaccharide blood group antigen. | | Descriptor: | FUCOLECTIN-RELATED PROTEIN, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Higgins, M.A, Whitworth, G.E, El Warry, N, Randriantsoa, M, Samain, E, Burke, R.D, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Recognition and Hydrolysis of Host Carbohydrate-Antigens by Streptococcus Pneumoniae Family 98 Glycoside Hydrolases.
J.Biol.Chem., 284, 2009
|
|
7MOY
 
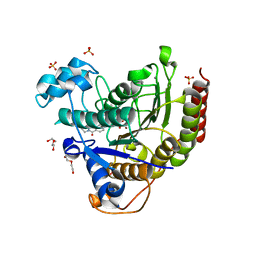 | | Structure of HDAC2 in complex with an inhibitor (compound 19) | | Descriptor: | (1S)-6-ethyl-N-{(1S)-1-[5-(2-ethyl-1-oxo-1,2-dihydroisoquinolin-6-yl)-1H-imidazol-2-yl]-7,7-dihydroxynonyl}-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7MOT
 
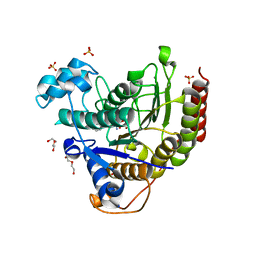 | | Structure of HDAC2 in complex with an inhibitor (compound 9) | | Descriptor: | 5-{(1S)-7,7-dihydroxy-1-[(1-methylazetidine-3-carbonyl)amino]nonyl}-2-phenyl-1H-imidazole-4-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7MOS
 
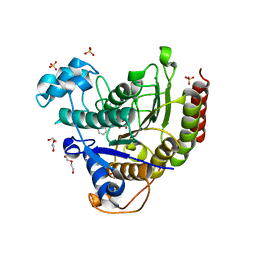 | | Structure of HDAC2 in complex with a macrocyclic inhibitor (compound 4) | | Descriptor: | (3S,18S,20aR)-18-(6,6-dihydroxyoctyl)-1,5,6,7,8,18,19,20a-octahydro-4H-14,17-epiminoazeto[1,2-g][1,7,10,13]benzoxatriazacycloheptadecin-20(2H)-one, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7MOX
 
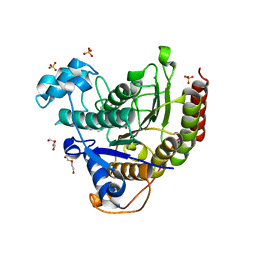 | | Structure of HDAC2 in complex with an inhibitor (compound 14) | | Descriptor: | (1S)-N-[(1S)-7,7-dihydroxy-1-{4-[(1R,4S)-1,2,3,4-tetrahydro-1,4-methanonaphthalen-6-yl]-1H-imidazol-2-yl}nonyl]-6-methyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
5IH6
 
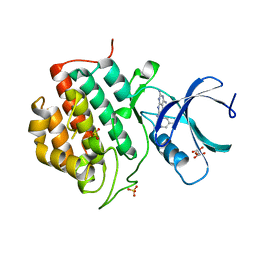 | | Human Casein Kinase 1 isoform delta (kinase domain) in complex with Epiblastin A derivative | | Descriptor: | 6-(3-bromophenyl)pteridine-2,4,7-triamine, Casein kinase I isoform delta, S,R MESO-TARTARIC ACID, ... | | Authors: | Ursu, A, Illich, D.J, Takemoto, Y, Porfetye, A.T, Zhang, M, Brockmeyer, A, Janning, P, Watanabe, N, Osada, H, Vetter, I.R, Ziegler, S, Schoeler, H.R, Waldmann, H. | | Deposit date: | 2016-02-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Epiblastin A Induces Reprogramming of Epiblast Stem Cells Into Embryonic Stem Cells by Inhibition of Casein Kinase 1.
Cell Chem Biol, 23, 2016
|
|
2XO8
 
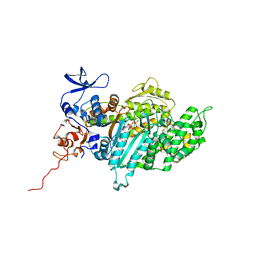 | | Crystal Structure of Myosin-2 in Complex with Tribromodichloropseudilin | | Descriptor: | 2,4-DICHLORO-6-(3,4,5-TRIBROMO-1H-PYRROL-2-YL)PHENOL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Preller, M, Chinthalapudi, K, Martin, R, Knoelker, H.J, Manstein, D.J. | | Deposit date: | 2010-08-10 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of Myosin ATPase Activity by Halogenated Pseudilins: A Structure-Activity Study.
J.Med.Chem., 54, 2011
|
|
1H2E
 
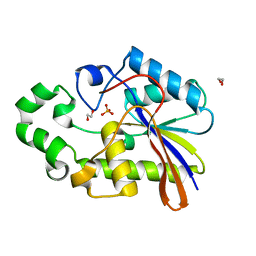 | | BACILLUS STEAROTHERMOPHILUS PHOE (previously known as yhfr) in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATASE, PHOSPHATE ION | | Authors: | Rigden, D.J, Littlejohn, J.E, Jedrzejas, M.J. | | Deposit date: | 2002-08-08 | | Release date: | 2002-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structures of Phosphate and Trivanadate Complexes of Bacillus Stearothermophilus Phosphatase Phoe: Structural and Functional Analysis in the Cofactor-Dependent Phosphoglycerate Mutase Superfamily
J.Mol.Biol., 325, 2003
|
|
2YA2
 
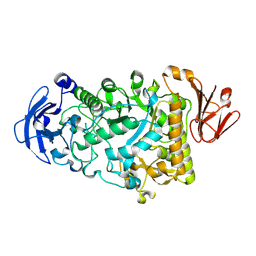 | | Catalytic Module of the Multi-modular glycogen-degrading pneumococcal virulence factor SpuA in complex with an inhibitor. | | Descriptor: | 1-DEOXYNOJIRIMYCIN, CALCIUM ION, PUTATIVE ALKALINE AMYLOPULLULANASE, ... | | Authors: | Lammerts van Bueren, A, Ficko-Blean, E, Pluvinage, B, Hehemann, J.H, Higgins, M.A, Deng, L, Ogunniyi, A.D, Stroeher, U.H, Warry, N.E, Burke, R.D, Czjzek, M, Paton, J.C, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The Conformation and Function of a Multimodular Glycogen-Degrading Pneumococcal Virulence Factor.
Structure, 19, 2011
|
|
2Y73
 
 | | THE NATIVE STRUCTURES OF SOLUBLE HUMAN PRIMARY AMINE OXIDASE AOC3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Elovaara, H, Kidron, H, Parkash, V, Nymalm, Y, Bligt, E, Ollikka, P, Smith, D.J, Pihlavisto, M, Salmi, M, Jalkanen, S, Salminen, T.A. | | Deposit date: | 2011-01-28 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Two Imidazole Binding Sites and Key Residues for Substrate Specificity in Human Primary Amine Oxidase Aoc3.
Biochemistry, 50, 2011
|
|
1H2F
 
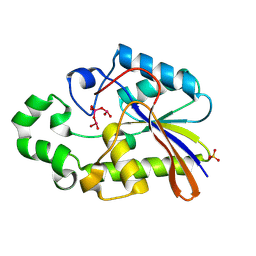 | |
7M7X
 
 | |
7MU6
 
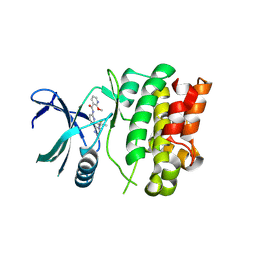 | | Ask1 bound to compound 28 | | Descriptor: | 2-methoxy-N-(6-{4-[(2S)-1,1,1-trifluoropropan-2-yl]-4H-1,2,4-triazol-3-yl}pyridin-2-yl)pyridine-3-carboxamide, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Chodaparambil, J.V, Marcotte, D.J. | | Deposit date: | 2021-05-14 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.165 Å) | | Cite: | Discovery of Potent, Selective, and Brain-Penetrant Apoptosis Signal-Regulating Kinase 1 (ASK1) Inhibitors that Modulate Brain Inflammation In Vivo.
J.Med.Chem., 64, 2021
|
|
5EUJ
 
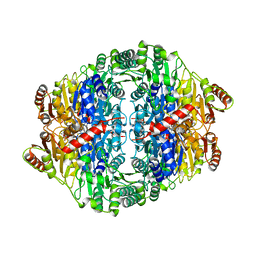 | | PYRUVATE DECARBOXYLASE FROM ZYMOBACTER PALMAE | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Pyruvate decarboxylase, ... | | Authors: | Buddrus, L, Crennell, S.J, Leak, D.J, Danson, M.J, Andrews, E.S.V, Arcus, V.L. | | Deposit date: | 2015-11-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of pyruvate decarboxylase from Zymobacter palmae.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
2VSA
 
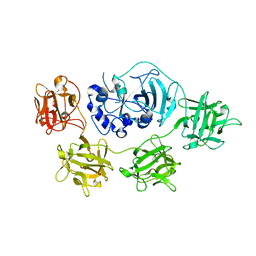 | | Structure and mode of action of a mosquitocidal holotoxin | | Descriptor: | MOSQUITOCIDAL TOXIN | | Authors: | Treiber, N, Reinert, D.J, Carpusca, I, Aktories, K, Schulz, G.E. | | Deposit date: | 2008-04-22 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure and Mode of Action of a Mosquitocidal Holotoxin
J.Mol.Biol., 381, 2008
|
|
5IWD
 
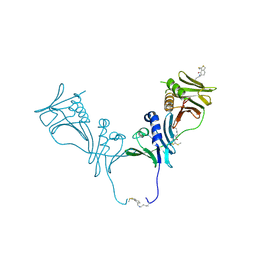 | | HCMV DNA polymerase subunit UL44 complex with a small molecule | | Descriptor: | 5-methylidene-3-(methylsulfanyl)-2-benzothiophen-4(5H)-one, DNA polymerase processivity factor | | Authors: | Chen, H, Coen, D.M, Hogle, J.M, Filman, D.J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A Small Covalent Allosteric Inhibitor of Human Cytomegalovirus DNA Polymerase Subunit Interactions.
ACS Infect Dis, 3, 2017
|
|
7MFT
 
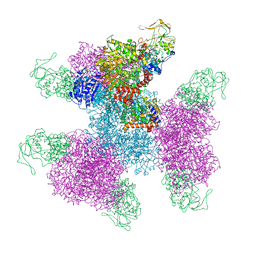 | | Glutamate synthase, glutamate dehydrogenase counter-enzyme complex (GudB6-GltA6-GltB6) | | Descriptor: | FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Jayaraman, V, Lee, D.J, Elad, N, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2021-04-11 | | Release date: | 2022-01-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A counter-enzyme complex regulates glutamate metabolism in Bacillus subtilis.
Nat.Chem.Biol., 18, 2022
|
|
7MFM
 
 | | Glutamate synthase, glutamate dehydrogenase counter-enzyme complex | | Descriptor: | FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Jayaraman, V, Lee, D.J, Elad, N, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2021-04-10 | | Release date: | 2022-01-05 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | A counter-enzyme complex regulates glutamate metabolism in Bacillus subtilis.
Nat.Chem.Biol., 18, 2022
|
|
5FJX
 
 | |
5IX2
 
 | | Crystal structure of mouse Morc3 ATPase-CW cassette in complex with AMPPNP and unmodified H3 peptide | | Descriptor: | MAGNESIUM ION, MORC family CW-type zinc finger protein 3, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Du, J, Patel, D.J. | | Deposit date: | 2016-03-23 | | Release date: | 2016-08-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2WP1
 
 | | Structure of Brdt bromodomain 2 bound to an acetylated histone H3 peptide | | Descriptor: | BROMODOMAIN TESTIS-SPECIFIC PROTEIN, HISTONE H3 | | Authors: | Moriniere, J, Rousseaux, S, Steuerwald, U, Soler-Lopez, M, Curtet, S, Vitte, A.-L, Govin, J, Gaucher, J, Sadoul, K, Hart, D.J, Krijgsveld, J, Khochbin, S, Mueller, C.W, Petosa, C. | | Deposit date: | 2009-08-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative Binding of Two Acetylation Marks on a Histone Tail by a Single Bromodomain.
Nature, 461, 2009
|
|
5JLH
 
 | | Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 2, ... | | Authors: | von der Ecken, J, Heissler, S.M, Pathan-Chhatbar, S, Manstein, D.J, Raunser, S. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-15 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution.
Nature, 534, 2016
|
|
7MKK
 
 | |
7MQQ
 
 | |
