1IDC
 
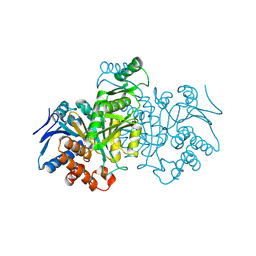 | | ISOCITRATE DEHYDROGENASE FROM E.COLI (MUTANT K230M), STEADY-STATE INTERMEDIATE COMPLEX DETERMINED BY LAUE CRYSTALLOGRAPHY | | Descriptor: | 2-OXALOSUCCINIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
1IDF
 
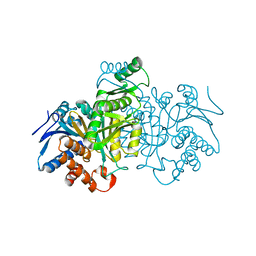 | | ISOCITRATE DEHYDROGENASE K230M MUTANT APO ENZYME | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
1IEB
 
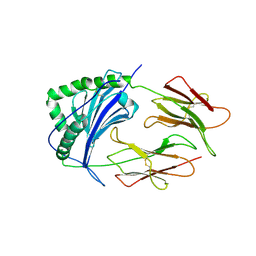 | | HISTOCOMPATIBILITY ANTIGEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS II I-EK, SULFATE ION | | Authors: | Fremont, D.H, Hendrickson, W.A, Marrack, P, Kappler, J. | | Deposit date: | 1996-04-05 | | Release date: | 1997-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of an MHC class II molecule with covalently bound single peptides.
Science, 272, 1996
|
|
6Y8L
 
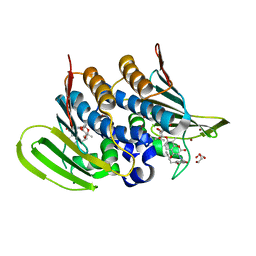 | | Mycobacterium thermoresistibile GyrB21 in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
3V52
 
 | | Structure of a monoclonal antibody complexed with its MHC-I antigen | | Descriptor: | 1,2-ETHANEDIOL, ANTI-MHC-I MONOCLONAL ANTIBODY, 64-3-7 H CHAIN, ... | | Authors: | Mage, M.G, Dolan, M.A, Wang, R, Boyd, L.F, Revilleza, M.J, Robinson, H, Natarajan, K, Myers, N.B, Hansen, T.H, Margulies, D.H. | | Deposit date: | 2011-12-15 | | Release date: | 2012-07-25 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | The Peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics.
J.Immunol., 189, 2012
|
|
8TQ7
 
 | | Crystal structure of Fab.34.2.12 in complex with MHC-I (H2-Dd) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab 34.2.12 Light Chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQ8
 
 | | Crystal structure of Fab.34.5.8 in complex with MHC-I (H2-Dd) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab.34.5.8 Heavy chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQ9
 
 | | Crystal structure of Fab.S19.8 in complex with MHC-I (H2-Dd) | | Descriptor: | Beta-2-microglobulin, Fab.S19.8 Heavy Chain, Fab.S19.8 Light Chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQA
 
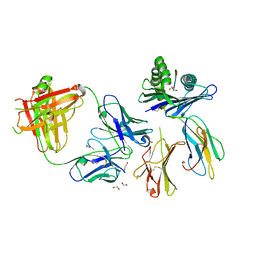 | | Crystal structure of Fab.28.14.8 in complex with MHC-I (H2-Db) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8R50
 
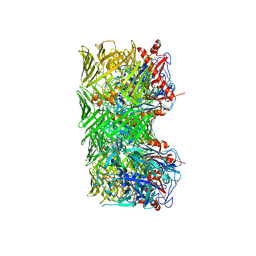 | | Mouse teneurin-3 compact dimer - A1B1 isoform | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Teneurin-3 | | Authors: | Gogou, C, Meijer, D.H. | | Deposit date: | 2023-11-15 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition.
Nat Commun, 15, 2024
|
|
8R51
 
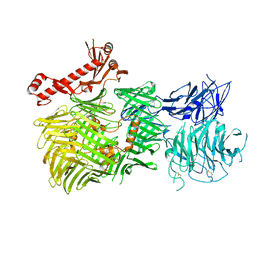 | | Mouse teneurin-3 non-compact subunit - A1B1 isoform | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Teneurin-3 | | Authors: | Gogou, C, Meijer, D.H. | | Deposit date: | 2023-11-15 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition.
Nat Commun, 15, 2024
|
|
8R54
 
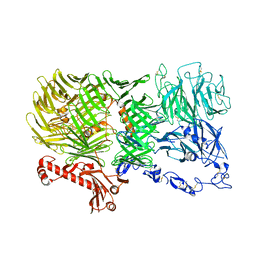 | | Mouse teneurin-3 non-compact subunit - A0B0 isoform | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Teneurin-3, ... | | Authors: | Gogou, C, Meijer, D.H. | | Deposit date: | 2023-11-15 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Alternative splicing controls teneurin-3 compact dimer formation for neuronal recognition.
Nat Commun, 15, 2024
|
|
1IDD
 
 | | ISOCITRATE DEHYDROGENASE Y160F MUTANT APO ENZYME | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Lee, M.E, Dyer, D.H, Klein, O.D, Bolduc, J.M, Stoddard, B.L, Koshland Junior, D.E. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
6Z83
 
 | | CK2 alpha bound to chemical probe SGC-CK2-1 | | Descriptor: | Casein kinase II subunit alpha, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Kraemer, A, Wells, C, Drewry, D.H, Pickett, J.E, Axtman, A.D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-02 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | Development of a potent and selective chemical probe for the pleiotropic kinase CK2.
Cell Chem Biol, 28, 2021
|
|
1J5L
 
 | | NMR STRUCTURE OF THE ISOLATED BETA_C DOMAIN OF LOBSTER METALLOTHIONEIN-1 | | Descriptor: | CADMIUM ION, METALLOTHIONEIN-1 | | Authors: | Munoz, A, Forsterling, F.H, Shaw III, C.F, Petering, D.H. | | Deposit date: | 2002-05-16 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the (113)Cd(3)beta domains from Homarus americanus metallothionein-1: hydrogen bonding and solvent accessibility of sulfur atoms
J.Biol.Inorg.Chem., 7, 2002
|
|
8XOA
 
 | | VMAT2 complex with MPP+ | | Descriptor: | 1-methyl-4-phenylpyridin-1-ium, Synaptic vesicular amine transporter,Synaptic vesicular amine transporter,transporterA | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-12-31 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | VMAT2 complex with MPP+
To Be Published
|
|
8XO9
 
 | | VMAT2 complex with noradrenaline in cytosol-facing state | | Descriptor: | Noradrenaline, Synaptic vesicular amine transporter,Synaptic vesicular amine transporter,transporter A | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-12-31 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | VMAT2 complex with noradrenaline in cytosol-facing state
To Be Published
|
|
8XOB
 
 | | VMAT2 protonated state | | Descriptor: | Synaptic vesicular amine transporter,transporter B | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-12-31 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | VMAT2 protonated state
To Be Published
|
|
1G93
 
 | | CRYSTAL STRUCTURE OF THE BOVINE CATALYTIC DOMAIN OF ALPHA-1,3-GALACTOSYLTRANSFERASE IN THE PRESENCE OF UDP-GALACTOSE | | Descriptor: | MANGANESE (II) ION, MERCURY (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Gastinel, L.N, Bignon, C, Misra, A.K, Hindsgaul, O, Shaper, J.H, Joziasse, D.H. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bovine alpha1,3-galactosyltransferase catalytic domain structure and its relationship with ABO histo-blood group and glycosphingolipid glycosyltransferases.
EMBO J., 20, 2001
|
|
3TSS
 
 | | TOXIC SHOCK SYNDROME TOXIN-1 TETRAMUTANT, P2(1) CRYSTAL FORM | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Prasad, G.S, Radhakrishnan, R, Mitchell, D.T, Earhart, C.A, Dinges, M.M, Cook, W.J, Schlivert, P.M, Ohlendorf, D.H. | | Deposit date: | 1996-12-11 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined structures of three crystal forms of toxic shock syndrome toxin-1 and of a tetramutant with reduced activity.
Protein Sci., 6, 1997
|
|
3TTK
 
 | | Crystal structure of apo-SpuD | | Descriptor: | Polyamine transport protein | | Authors: | Wu, D.H, Lim, S.C, Song, H.W. | | Deposit date: | 2011-09-14 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural Basis of Substrate Binding Specificity Revealed by the Crystal Structures of Polyamine Receptors SpuD and SpuE from Pseudomonas aeruginosa
J.Mol.Biol., 416, 2012
|
|
1GLO
 
 | | Crystal Structure of Cys25Ser mutant of human cathepsin S | | Descriptor: | CATHEPSIN S | | Authors: | Turkenburg, J.P, Lamers, M.B.A.C, Brzozowski, A.M, Wright, L.M, Hubbard, R.E, Sturt, S.L, Williams, D.H. | | Deposit date: | 2001-08-31 | | Release date: | 2002-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a Cys25->Ser Mutant of Human Cathepsin Cathepsin S
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1J5M
 
 | | SOLUTION STRUCTURE OF THE SYNTHETIC 113CD_3 BETA_N DOMAIN OF LOBSTER METALLOTHIONEIN-1 | | Descriptor: | CADMIUM ION, METALLOTHIONEIN-1 | | Authors: | Munoz, A, Forsterling, F.H, Shaw III, C.F, Petering, D.H. | | Deposit date: | 2002-05-16 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the (113)Cd(3)beta domains from Homarus americanus metallothionein-1: hydrogen bonding and solvent accessibility of sulfur atoms
J.Biol.Inorg.Chem., 7, 2002
|
|
1G4S
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, THIAMIN PHOSPHATE, ... | | Authors: | Peapus, D.H, Chiu, H.-J, Campobasso, N, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-10-27 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase.
Biochemistry, 40, 2001
|
|
3VW7
 
 | | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Proteinase-activated receptor 1, ... | | Authors: | Zhang, C, Srinivasan, Y, Arlow, D.H, Fung, J.J, Palmer, D, Zheng, Y, Green, H.F, Pandey, A, Dror, R.O, Shaw, D.E, Weis, W.I, Coughlin, S.R, Kobilka, B.K. | | Deposit date: | 2012-08-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structure of human protease-activated receptor 1
Nature, 492, 2012
|
|
