1R0D
 
 | |
1QQS
 
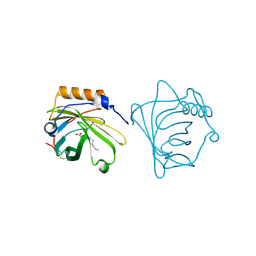 | | NEUTROPHIL GELATINASE ASSOCIATED LIPOCALIN HOMODIMER | | 分子名称: | DECANOIC ACID, NEUTROPHIL GELATINASE, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Goetz, D.H, Willie, S.T, Armen, R, Bratt, T, Borregaard, N, Strong, R.K. | | 登録日 | 1999-06-07 | | 公開日 | 2000-04-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Ligand preference inferred from the structure of neutrophil gelatinase associated lipocalin
Biochemistry, 39, 2000
|
|
1QTP
 
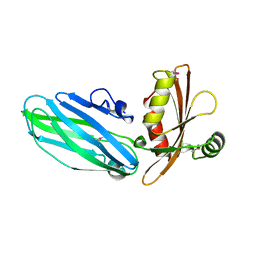 | |
1QTF
 
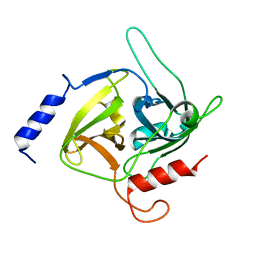 | | CRYSTAL STRUCTURE OF EXFOLIATIVE TOXIN B | | 分子名称: | EXFOLIATIVE TOXIN B | | 著者 | Vath, G.M, Earhart, C.A, Monie, D.D, Schlievert, P.M, Ohlendorf, D.H. | | 登録日 | 1999-06-27 | | 公開日 | 1999-08-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure of exfoliative toxin B: a superantigen with enzymatic activity.
Biochemistry, 38, 1999
|
|
5CWV
 
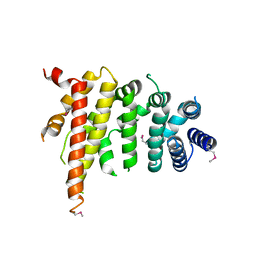 | | Crystal structure of Chaetomium thermophilum Nup192 TAIL domain | | 分子名称: | Nucleoporin NUP192 | | 著者 | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | 登録日 | 2015-07-28 | | 公開日 | 2015-09-16 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (3.155 Å) | | 主引用文献 | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
1IY7
 
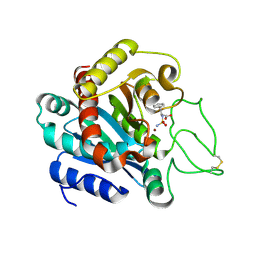 | | Crystal Structure of CPA and sulfamide-based inhibitor complex | | 分子名称: | Carboxypeptidase A, PHENYLALANINE-N-SULFONAMIDE, ZINC ION | | 著者 | Kim, S.J, Woo, J.R, Park, J.D, Kim, D.H, Ryu, S.E. | | 登録日 | 2002-07-24 | | 公開日 | 2003-01-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Sulfamide-Based Inhibitors for Carboxypeptidase A. Novel Type Transition State Analogue Inhibitors for Zinc Proteases
J.Med.Chem., 45, 2002
|
|
6V4N
 
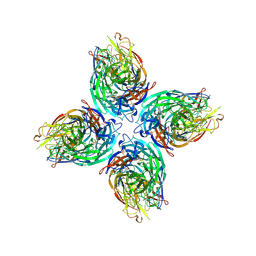 | |
6V4O
 
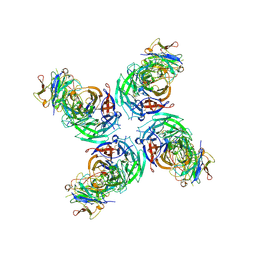 | |
1DMQ
 
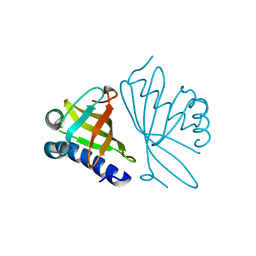 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | 分子名称: | STEROID DELTA-ISOMERASE | | 著者 | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | 登録日 | 1999-12-14 | | 公開日 | 2000-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1IEA
 
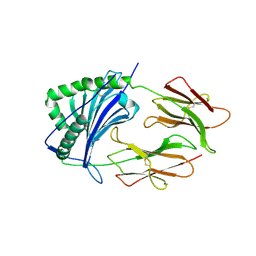 | | HISTOCOMPATIBILITY ANTIGEN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS II I-EK | | 著者 | Fremont, D.H, Hendrickson, W.A, Marrack, P, Kappler, J. | | 登録日 | 1996-04-05 | | 公開日 | 1997-06-05 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of an MHC class II molecule with covalently bound single peptides.
Science, 272, 1996
|
|
1IE6
 
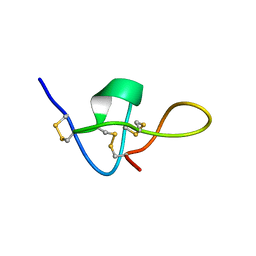 | | SOLUTION STRUCTURE OF IMPERATOXIN A | | 分子名称: | IMPERATOXIN A | | 著者 | Lee, C.W, Takeuchi, K, Takahashi, H, Sato, K, Shimada, I, Kim, D.H, Kim, J.I. | | 登録日 | 2001-04-07 | | 公開日 | 2003-06-10 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular basis of the high-affinity activation of type 1 ryanodine receptors by imperatoxin A.
Biochem.J., 377, 2004
|
|
6VBK
 
 | | Crystal structure of N-terminal domain of Mycobacterium tuberculosis complex Lon protease | | 分子名称: | GLYCEROL, Lon211 | | 著者 | Bi, F.K, Chen, C, Chen, X.Y, Guo, C.Y, Lin, D.H. | | 登録日 | 2019-12-19 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the N domain of Lon protease from Mycobacterium avium complex.
Protein Sci., 28, 2019
|
|
6VET
 
 | | Human insulin analog: [GluB10,HisA8,ArgA9,TyrB20]-DOI | | 分子名称: | Insulin A chain, Insulin B chain | | 著者 | Menting, J.G, Chou, D.H.-C, Lawrence, M.C, Xiong, X. | | 登録日 | 2020-01-02 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | A structurally minimized yet fully active insulin based on cone-snail venom insulin principles.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VER
 
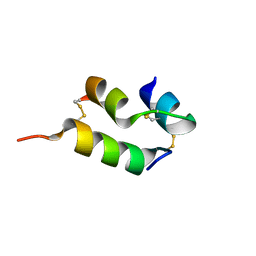 | | Human insulin analog: [GluB10,TyrB20]-DOI | | 分子名称: | Insulin A chain, Insulin B chain | | 著者 | Menting, J.G, Chou, D.H.-C, Lawrence, M.C, Xiong, X. | | 登録日 | 2020-01-02 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.047 Å) | | 主引用文献 | Mini-Ins: A minimal, bioactive insulin analog with alternative binding modes
not published
|
|
1S0L
 
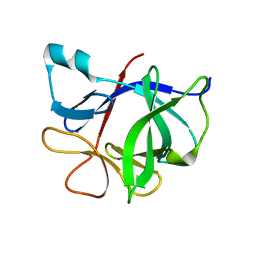 | |
1IDC
 
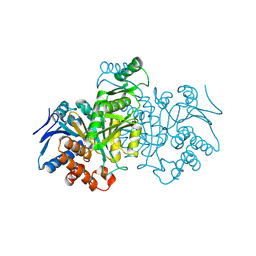 | | ISOCITRATE DEHYDROGENASE FROM E.COLI (MUTANT K230M), STEADY-STATE INTERMEDIATE COMPLEX DETERMINED BY LAUE CRYSTALLOGRAPHY | | 分子名称: | 2-OXALOSUCCINIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION | | 著者 | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | 登録日 | 1995-01-18 | | 公開日 | 1996-03-08 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
1S7D
 
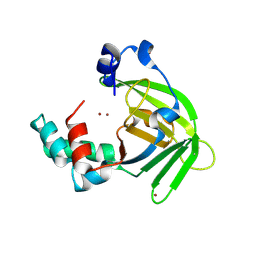 | |
1S7C
 
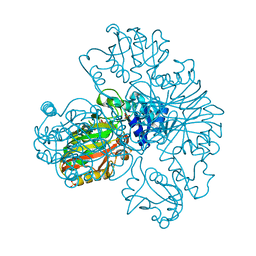 | | Crystal structure of MES buffer bound form of glyceraldehyde 3-phosphate dehydrogenase from Escherichia coli | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glyceraldehyde 3-phosphate dehydrogenase A, SULFATE ION | | 著者 | Shin, D.H, Thor, J, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 2004-01-29 | | 公開日 | 2004-08-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Crystal structure of MES buffer bound form of glyceraldehyde 3-phosphate dehydrogenase from Escherichia coli
To be Published
|
|
1IDD
 
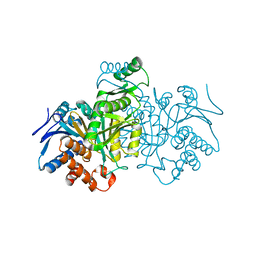 | | ISOCITRATE DEHYDROGENASE Y160F MUTANT APO ENZYME | | 分子名称: | ISOCITRATE DEHYDROGENASE | | 著者 | Lee, M.E, Dyer, D.H, Klein, O.D, Bolduc, J.M, Stoddard, B.L, Koshland Junior, D.E. | | 登録日 | 1995-01-18 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
1QES
 
 | | TANDEM GU MISMATCHES IN RNA, NMR, 30 STRUCTURES | | 分子名称: | RNA (5'-R(*GP*GP*AP*GP*UP*UP*CP*C)-3') | | 著者 | Mcdowell, J.A, He, L, Chen, X, Turner, D.H. | | 登録日 | 1997-03-04 | | 公開日 | 1997-06-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Investigation of the structural basis for thermodynamic stabilities of tandem GU wobble pairs: NMR structures of (rGGAGUUCC)2 and (rGGAUGUCC)2.
Biochemistry, 36, 1997
|
|
1EKD
 
 | | NMR AND MOLECULAR MODELING REVEAL THAT DIFFERENT HYDROGEN BONDING PATTERNS ARE POSSIBLE FOR GU PAIRS: ONE HYDROGEN BOND FOR EACH GU PAIR IN R(GGCGUGCC)2 AND TWO FOR EACH GU PAIR IN R(GAGUGCUC)2 | | 分子名称: | RNA (5'-R(*GP*GP*CP*GP*UP*GP*CP*C)-3') | | 著者 | Chen, X, McDowell, J.A, Kierzek, R, Krugh, T.R, Turner, D.H. | | 登録日 | 2000-03-07 | | 公開日 | 2000-11-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2).
Biochemistry, 39, 2000
|
|
8SQN
 
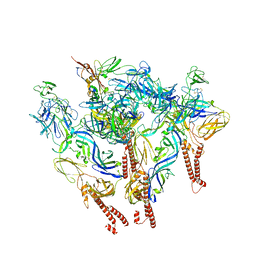 | | CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DuD1MoD2 chimeric MXRA8, E1 envelope glycoprotein, ... | | 著者 | Zimmerman, M.I, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2023-05-04 | | 公開日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.89 Å) | | 主引用文献 | Vertebrate-class-specific binding modes of the alphavirus receptor MXRA8.
Cell, 186, 2023
|
|
1J5L
 
 | | NMR STRUCTURE OF THE ISOLATED BETA_C DOMAIN OF LOBSTER METALLOTHIONEIN-1 | | 分子名称: | CADMIUM ION, METALLOTHIONEIN-1 | | 著者 | Munoz, A, Forsterling, F.H, Shaw III, C.F, Petering, D.H. | | 登録日 | 2002-05-16 | | 公開日 | 2002-05-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the (113)Cd(3)beta domains from Homarus americanus metallothionein-1: hydrogen bonding and solvent accessibility of sulfur atoms
J.Biol.Inorg.Chem., 7, 2002
|
|
6VCC
 
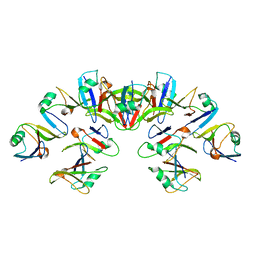 | | Cryo-EM structure of the Dvl2 DIX filament | | 分子名称: | Segment polarity protein dishevelled homolog DVL-2 | | 著者 | Enos, M, Kan, W, Muennich, S, Chen, D.H, Skiniotis, G, Weis, W.I. | | 登録日 | 2019-12-20 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Limited Dishevelled/Axin oligomerization determines efficiency of Wnt/ beta-catenin signal transduction.
Elife, 9, 2020
|
|
1EYH
 
 | |
