7UWY
 
 | | NMR solution structure of the De novo designed small beta-barrel protein 29_bp_sh3 | | Descriptor: | De novo designed small beta-barrel protein 29_bp_sh3 | | Authors: | Peterson, F.C, Kim, D.E, Jensen, D.R, Saleem, A, Chow, C.M, Volkman, B.F, Baker, D. | | Deposit date: | 2022-05-04 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of small beta barrel proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7UWZ
 
 | | NMR solution structure of the De novo designed small beta-barrel protein 33_bp_sh3 | | Descriptor: | De novo designed small beta-barrel protein 33_bp_sh3 | | Authors: | Peterson, F.C, Kim, D.E, Jensen, D.R, Saleem, A, Chow, C.M, Volkman, B.F, Baker, D. | | Deposit date: | 2022-05-04 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of small beta barrel proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7UKV
 
 | | Wild type EGFR in complex with Lazertinib (YH25448) | | Descriptor: | Epidermal growth factor receptor, N-[5-{[(4P)-4-{4-[(dimethylamino)methyl]-3-phenyl-1H-pyrazol-1-yl}pyrimidin-2-yl]amino}-4-methoxy-2-(morpholin-4-yl)phenyl]propanamide | | Authors: | Beyett, T.S, Pham, C, Eck, M.J, Heppner, D.E. | | Deposit date: | 2022-04-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Inhibition of Mutant EGFR with Lazertinib (YH25448).
Acs Med.Chem.Lett., 13, 2022
|
|
7UKW
 
 | | EGFR(T790M/V948R) in complex with Lazertinib (YH25448) | | Descriptor: | Epidermal growth factor receptor, N-[5-{[(4P)-4-{4-[(dimethylamino)methyl]-3-phenyl-1H-pyrazol-1-yl}pyrimidin-2-yl]amino}-4-methoxy-2-(morpholin-4-yl)phenyl]propanamide | | Authors: | Pham, C.D, Heppner, D.E. | | Deposit date: | 2022-04-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Inhibition of Mutant EGFR with Lazertinib (YH25448).
Acs Med.Chem.Lett., 13, 2022
|
|
7ICD
 
 | | REGULATION OF AN ENZYME BY PHOSPHORYLATION AT THE ACTIVE SITE | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Hurley, J.H, Dean, A.M, Sohl, J.L, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1990-05-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulation of an enzyme by phosphorylation at the active site.
Science, 249, 1990
|
|
4ICD
 
 | | REGULATION OF ISOCITRATE DEHYDROGENASE BY PHOSPHORYLATION INVOLVES NO LONG-RANGE CONFORMATIONAL CHANGE IN THE FREE ENZYME | | Descriptor: | PHOSPHORYLATED ISOCITRATE DEHYDROGENASE | | Authors: | Hurley, J.H, Dean, A.M, Thorsness, P.E, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1989-12-28 | | Release date: | 1991-01-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of isocitrate dehydrogenase by phosphorylation involves no long-range conformational change in the free enzyme.
J.Biol.Chem., 265, 1990
|
|
7AB5
 
 | |
7AB4
 
 | |
7AB3
 
 | |
7B0Y
 
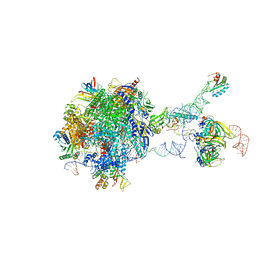 | | Structure of a transcribing RNA polymerase II-U1 snRNP complex | | Descriptor: | 145-nt RNA, DNA-directed RNA polymerase II subunit D, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Zhang, S, Aibara, S, Vos, S.M, Agafonov, D.E, Luehrmann, R, Cramer, P. | | Deposit date: | 2020-11-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a transcribing RNA polymerase II-U1 snRNP complex.
Science, 371, 2021
|
|
7YWS
 
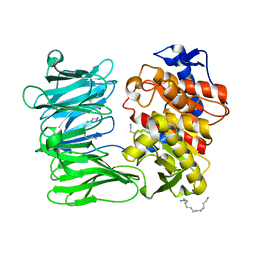 | | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 3 spermine molecules at 1.7 A resolution | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Elucidation of the Conformational Transition of Oligopeptidase B by an Integrative Approach Based on the Combination of X-ray, SAXS, and Essential Dynamics Sampling Simulation
Crystals, 12, 2022
|
|
7YX7
 
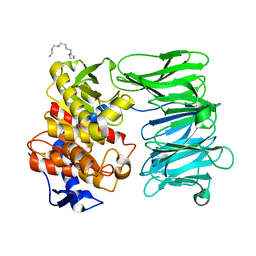 | | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 1 spermine molecule at 1.72 A resolution | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-02-15 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Elucidation of the Conformational Transition of Oligopeptidase B by an Integrative Approach Based on the Combination of X-ray, SAXS, and Essential Dynamics Sampling Simulation
Crystals, 12, 2022
|
|
7YWP
 
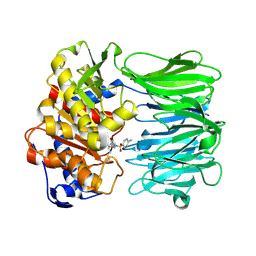 | | Closed conformation of Oligopeptidase B from Serratia proteomaculans with covalently bound TCK | | Descriptor: | N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide, Oligopeptidase B | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Inhibitor-Bound Bacterial Oligopeptidase B in the Closed State: Similarity and Difference between Protozoan and Bacterial Enzymes.
Int J Mol Sci, 24, 2023
|
|
7YWZ
 
 | | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 4 spermine molecules at 1.75 A resolution | | Descriptor: | GLYCEROL, Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-02-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 4 spermine molecules at 1.75 A resolution
To Be Published
|
|
7ZJZ
 
 | | catalytically non active S532A mutant of oligopeptidase B from S. proteomaculans | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-04-12 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Elucidation of the Conformational Transition of Oligopeptidase B by an Integrative Approach Based on the Combination of X-ray, SAXS, and Essential Dynamics Sampling Simulation
Crystals, 12, 2022
|
|
6ZN8
 
 | | Crystal structure of the H. influenzae VapXD toxin-antitoxin complex | | Descriptor: | Endoribonuclease VapD, VapX | | Authors: | Bertelsen, M.B, Senissar, M, Nielsen, M.H, Bisiak, F, Cunha, M.V, Molinaro, A.L, Daines, D.A, Brodersen, D.E. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Structural Basis for Toxin Inhibition in the VapXD Toxin-Antitoxin System.
Structure, 29, 2021
|
|
4V4T
 
 | | Crystal structure of the whole ribosomal complex with a stop codon in the A-site. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.46 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
3LTG
 
 | |
3LGK
 
 | | D99N Epi-isozizaene synthase | | Descriptor: | Epi-isozizaene synthase, SULFATE ION | | Authors: | Aaron, J.A, Lin, X, Cane, D.E, Christianson, D.W. | | Deposit date: | 2010-01-20 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structure of Epi-Isozizaene Synthase from Streptomyces coelicolor A3(2), a Platform for New Terpenoid Cyclization Templates
Biochemistry, 49, 2010
|
|
3LG5
 
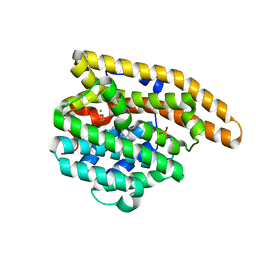 | | F198A Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, ... | | Authors: | Aaron, J.A, Lin, X, Cane, D.E, Christianson, D.W. | | Deposit date: | 2010-01-19 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structure of Epi-Isozizaene Synthase from Streptomyces coelicolor A3(2), a Platform for New Terpenoid Cyclization Templates
Biochemistry, 49, 2010
|
|
4XB6
 
 | | Structure of the E. coli C-P lyase core complex | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, ... | | Authors: | Brodersen, D.E. | | Deposit date: | 2014-12-16 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the bacterial carbon-phosphorus lyase machinery.
Nature, 525, 2015
|
|
3LTF
 
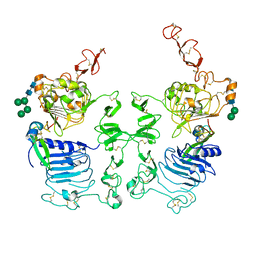 | | Crystal Structure of the Drosophila Epidermal Growth Factor Receptor ectodomain in complex with Spitz | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Alvarado, D, Klein, D.E, Lemmon, M.A. | | Deposit date: | 2010-02-15 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for negative cooperativity in growth factor binding to an EGF receptor.
Cell(Cambridge,Mass.), 142, 2010
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQW
 
 | | X-ray structure analysis of xylanase-N44E with MES at pH6.0 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XPV
 
 | | Neutron and X-ray structure analysis of xylanase: N44D at pH6 | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
