7KWY
 
 | | X-ray Crystal Structure of PlyCB Mutant R66K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
7NE7
 
 | | oligopeptidase B from S. proteomaculans with modified hinge region in complex with N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide | | Descriptor: | N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide, Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | Deposit date: | 2021-02-03 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of N alpha-p-tosyl-lysyl Chloromethylketone-Bound Oligopeptidase B from Serratia Proteamaculans Revealed a New Type of Inhibitor Binding
Crystals, 11, 2021
|
|
7NE4
 
 | | E125A mutant of oligopeptidase B from S. proteomaculans with modified hinge region | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | First Crystal Structure of Bacterial Oligopeptidase B in an Intermediate State: The Roles of the Hinge Region Modification and Spermine.
Biology (Basel), 10, 2021
|
|
4AY5
 
 | | Human O-GlcNAc transferase (OGT) in complex with UDP and glycopeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GTAB1TIDE, UDP-N-ACETYLGLUCOSAMINE--PEPTIDE N-ACETYLGLUCOSAMINYL TRANSFERASE 110 KDA SUBUNIT, ... | | Authors: | Schimpl, M, Zheng, X, Blair, D.E, Schuettelkopf, A.W, Navratilova, I, Aristotelous, T, Ferenbach, A.T, Macnaughtan, M.A, Borodkin, V.S, van Aalten, D.M.F. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | O-Glcnac Transferase Invokes Nucleotide Sugar Pyrophosphate Participation in Catalysis
Nat.Chem.Biol., 8, 2012
|
|
7MK7
 
 | | Augmentor domain of augmentor-beta | | Descriptor: | ALK and LTK ligand 1,Maltodextrin-binding protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Krimmer, S.G, Reshetnyak, A.V, Puleo, D.E, Schlessinger, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42815185 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
7M6C
 
 | | Crystal structure of PLA2 from snake venom of peruvian Bothrops atrox | | Descriptor: | Basic phospholipase A2, LAURIC ACID | | Authors: | Leonardo, D.A, Chojnowski, G, Simpkin, A, Seifert-Davila, W, Vivas-Ruiz, D.E, Keegan, R, Rigden, D. | | Deposit date: | 2021-03-25 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | findMySequence: a neural-network-based approach for identification of unknown proteins in X-ray crystallography and cryo-EM
Iucrj, 9, 2022
|
|
7MBJ
 
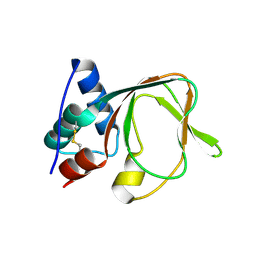 | |
7MYV
 
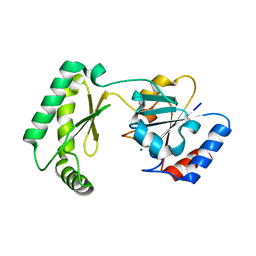 | | Plasmodium falciparum HAD5/PMM | | Descriptor: | MAGNESIUM ION, Phosphomannomutase | | Authors: | Frasse, P.M, Odom John, A.R, Goldberg, D.E. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Enzymatic and structural characterization of HAD5, an essential phosphomannomutase of malaria-causing parasites.
J.Biol.Chem., 298, 2022
|
|
3ZBW
 
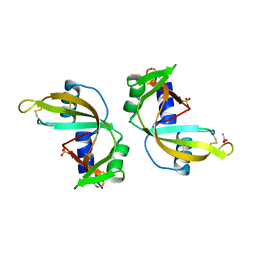 | | Crystal Structure of murine Angiogenin-3 | | Descriptor: | ACETIC ACID, ANGIOGENIN-3, SULFATE ION, ... | | Authors: | Iyer, S, Holloway, D.E, Acharya, K.R. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal Structures of Murine Angiogenin-2 and -3 - Probing 'Structure - Function' Relationships Amongst Angiogenin Homologues.
FEBS J., 280, 2013
|
|
4B3B
 
 | | Humanised monomeric RadA in complex with FHTA tetrapeptide | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, FHTA TETRAPEPTIDE, PHOSPHATE ION | | Authors: | Scott, D.E, Ehebauer, M.T, Pukala, T, Marsh, M, Blundell, T.L, Venkitaraman, A.R, Abell, C, Hyvonen, M. | | Deposit date: | 2012-07-23 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.193 Å) | | Cite: | Using a Fragment-Based Approach to Target Protein-Protein Interactions.
Chembiochem, 14, 2013
|
|
4B3C
 
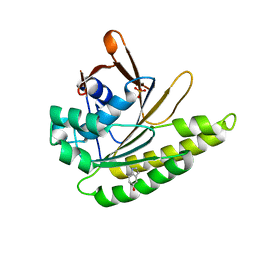 | | Humanised monomeric RadA in complex with 5-hydroxy indole | | Descriptor: | 1H-indol-5-ol, DNA REPAIR AND RECOMBINATION PROTEIN RADA, PHOSPHATE ION | | Authors: | Scott, D.E, Ehebauer, M.T, Pukala, T, Marsh, M, Blundell, T.L, Venkitaraman, A.R, Abell, C, Hyvonen, M. | | Deposit date: | 2012-07-23 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Using a Fragment-Based Approach to Target Protein-Protein Interactions.
Chembiochem, 14, 2013
|
|
4BC7
 
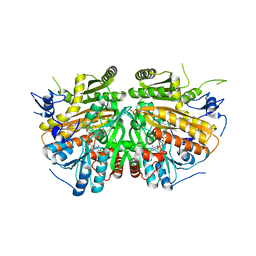 | | MAMMALIAN ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE: Arg419His mutant | | Descriptor: | 1-DODECANOL, ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, PEROXISOMAL, ... | | Authors: | Nenci, S, Piano, V, Rosati, S, Aliverti, A, Pandini, V, Fraaije, M.W, Heck, A.J.R, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Precursor of Ether Phospholipids is Synthesized by a Flavoenzyme Through Covalent Catalysis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4B32
 
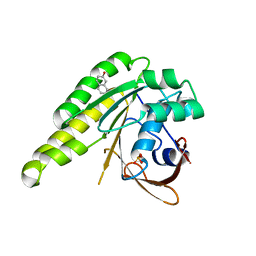 | | Humanised monomeric RadA in complex with napht-1-ol | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, PHOSPHATE ION, naphthalen-2-ol | | Authors: | Scott, D.E, Ehebauer, M.T, Pukala, T, Marsh, M, Blundell, T.L, Venkitaraman, A.R, Abell, C, Hyvonen, M. | | Deposit date: | 2012-07-20 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Using a Fragment-Based Approach to Target Protein-Protein Interactions.
Chembiochem, 14, 2013
|
|
4BC9
 
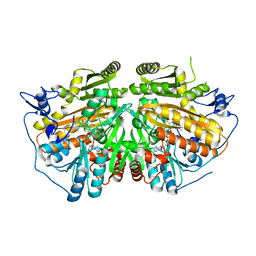 | | MAMMALIAN ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE: WILD-TYPE, ADDUCT WITH CYANOETHYL | | Descriptor: | ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, PEROXISOMAL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nenci, S, Piano, V, Rosati, S, Aliverti, A, Pandini, V, Fraaije, M.W, Heck, A.J.R, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Precursor of Ether Phospholipids is Synthesized by a Flavoenzyme Through Covalent Catalysis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4BCL
 
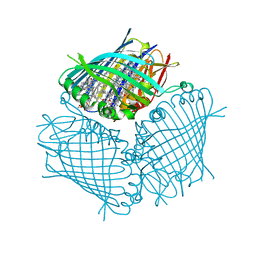 | | FMO protein from Prosthecochloris aestuarii 2K at Room Temperature | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOCHLOROPHYLL A PROTEIN | | Authors: | Tronrud, D.E, Matthews, B.W. | | Deposit date: | 1998-04-17 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refinement of the Structure of a Water-Soluble Antenna Complex from Green Photosynthetic Bacteria by Incorporation of the Chemically Determined Amino Acid Sequence
Photosynthetic Reaction Center, 1, 1993
|
|
1GK5
 
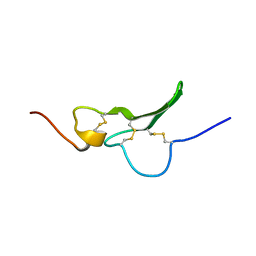 | | Solution Structure the mEGF/TGFalpha44-50 chimeric growth factor | | Descriptor: | Pro-epidermal growth factor,Protransforming growth factor alpha | | Authors: | Chamberlin, S.G, Brennan, L, Puddicombe, S.M, Davies, D.E, Turner, D.L. | | Deposit date: | 2001-08-08 | | Release date: | 2002-08-08 | | Last modified: | 2018-03-28 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Megf/Tgfalpha44-50 Chimeric Growth Factor.
Eur.J.Biochem., 268, 2001
|
|
2M20
 
 | | EGFR transmembrane - juxtamembrane (TM-JM) segment in bicelles: MD guided NMR refined structure. | | Descriptor: | Epidermal growth factor receptor | | Authors: | Endres, N.F, Das, R, Smith, A, Arkhipov, A, Kovacs, E, Huang, Y, Pelton, J.G, Shan, Y, Shaw, D.E, Wemmer, D.E, Groves, J.T, Kuriyan, J. | | Deposit date: | 2012-12-11 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational Coupling across the Plasma Membrane in Activation of the EGF Receptor.
Cell(Cambridge,Mass.), 152, 2013
|
|
1AC4
 
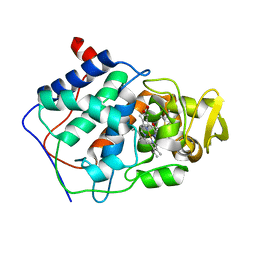 | | VARIATION IN THE STRENGTH OF A CH TO O HYDROGEN BOND IN AN ARTIFICIAL PROTEIN CAVITY (2,3,4-TRIMETHYL-1,3-THIAZOLE) | | Descriptor: | 2,3,4-TRIMETHYL-1,3-THIAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Bunte, S.W, Rosenfeld, R, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-12 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variation in strength of an unconventional C-H to O hydrogen bond in an engineered protein cavity
J.Am.Chem.Soc., 119, 1997
|
|
108D
 
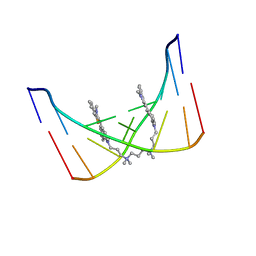 | | THE SOLUTION STRUCTURE OF A DNA COMPLEX WITH THE FLUORESCENT BIS INTERCALATOR TOTO DETERMINED BY NMR SPECTROSCOPY | | Descriptor: | 1,1-(4,4,8,8-TETRAMETHYL-4,8-DIAZAUNDECAMETHYLENE)-BIS-4-3-METHYL-2,3-DIHYDRO-(BENZO-1,3-THIAZOLE)-2-METHYLIDENE)-QUINOLINIUM, DNA (5'-D(*CP*GP*CP*TP*AP*GP*CP*G)-3') | | Authors: | Spielmann, H.P, Wemmer, D.E, Jacobsen, J.P. | | Deposit date: | 1995-01-31 | | Release date: | 1995-06-03 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA complex with the fluorescent bis-intercalator TOTO determined by NMR spectroscopy.
Biochemistry, 34, 1995
|
|
1AEE
 
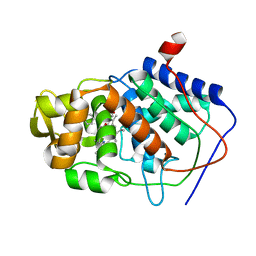 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (ANILINE) | | Descriptor: | ANILINE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-24 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
1AEU
 
 | | SPECIFICITY OF LIGAND BINDING IN A POLAR CAVITY OF CYTOCHROME C PEROXIDASE (2-METHYLIMIDAZOLE) | | Descriptor: | 2-METHYLIMIDAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A ligand-gated, hinged loop rearrangement opens a channel to a buried artificial protein cavity.
Nat.Struct.Biol., 3, 1996
|
|
1AJR
 
 | | REFINEMENT AND COMPARISON OF THE CRYSTAL STRUCTURES OF PIG CYTOSOLIC ASPARTATE AMINOTRANSFERASE AND ITS COMPLEX WITH 2-METHYLASPARTATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE | | Authors: | Rhee, S, Silva, M.M, Hyde, C.C, Rogers, P.H, Metzler, C.M, Metzler, D.E, Arnone, A. | | Deposit date: | 1997-05-08 | | Release date: | 1997-08-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Refinement and comparisons of the crystal structures of pig cytosolic aspartate aminotransferase and its complex with 2-methylaspartate.
J.Biol.Chem., 272, 1997
|
|
1AEK
 
 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (INDOLINE) | | Descriptor: | CYTOCHROME C PEROXIDASE, INDOLINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
1AA4
 
 | | SPECIFICITY OF LIGAND BINDING IN A BURIED POLAR CAVITY OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-01-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A ligand-gated, hinged loop rearrangement opens a channel to a buried artificial protein cavity.
Nat.Struct.Biol., 3, 1996
|
|
1AEN
 
 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (2-AMINO-5-METHYLTHIAZOLE) | | Descriptor: | 2-AMINO-5-METHYLTHIAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Bunte, S.W, Rosenfeld, R, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
