5IP4
 
 | | X-RAY STRUCTURE OF THE C-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin, XA4551 NANOBODY AGAINST C-DCX | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5FIP
 
 | | Discovery and characterization of a novel thermostable and highly halotolerant GH5 cellulase from an Icelandic hot spring isolate | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CHLORIDE ION, ... | | Authors: | Zarafeta, D, Kissas, D, Sayer, C, Gudbergsdottir, S.R, Ladoukakis, E, Isupov, M.N, Chatziioannou, A, Peng, X, Littlechild, J.A, Skretas, G, Kolisis, F.N. | | Deposit date: | 2015-10-01 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and Characterization of a Thermostable and Highly Halotolerant Gh5 Cellulase from an Icelandic Hot Spring Isolate.
Plos One, 11, 2016
|
|
6SNE
 
 | | crystal structure of LN01 Fab in complex with an HIV-1 gp41 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, LN01 heavy chain, ... | | Authors: | Caillat, C, Pinto, D, Corti, D, Fenwick, C, Pantaleo, G, Weissenhorn, W. | | Deposit date: | 2019-08-23 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Basis for Broad HIV-1 Neutralization by the MPER-Specific Human Broadly Neutralizing Antibody LN01.
Cell Host Microbe, 26, 2019
|
|
1RVA
 
 | |
1RNZ
 
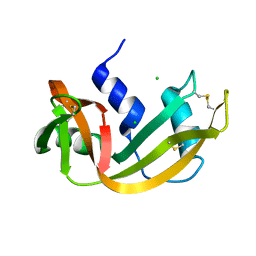 | | RIBONUCLEASE A CRYSTALLIZED FROM 2.5M SODIUM CHLORIDE, 3.3M SODIUM FORMATE | | Descriptor: | CHLORIDE ION, RIBONUCLEASE A | | Authors: | Fedorov, A.A, Josph-Mccarthy, D, Fedorov, E.V, Sirakova, D, Graf, I, Almo, S.C. | | Deposit date: | 1996-11-08 | | Release date: | 1997-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ionic interactions in crystalline bovine pancreatic ribonuclease A.
Biochemistry, 35, 1996
|
|
1S0V
 
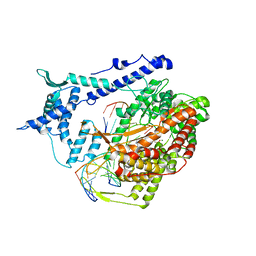 | | Structural basis for substrate selection by T7 RNA polymerase | | Descriptor: | 5'-D(*G*GP*GP*AP*AP*TP*CP*GP*AP*TP*AP*TP*CP*GP*CP*CP*GP*C)-3', 5'-D(*GP*TP*CP*GP*AP*TP*TP*CP*CP*C)-3', 5'-R(*AP*AP*CP*U*GP*CP*GP*GP*CP*GP*AP*U)-3', ... | | Authors: | Temiakov, D, Patlan, V, Anikin, M, McAllister, W.T, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-05 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for substrate selection by t7 RNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
5FHY
 
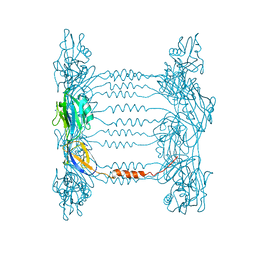 | | Crystal structure of FliD (HAP2) from Pseudomonas aeruginosa PAO1 | | Descriptor: | B-type flagellar hook-associated protein 2, SODIUM ION | | Authors: | Postel, S, Bonsor, D, Diederichs, K, Sundberg, E.J. | | Deposit date: | 2015-12-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Bacterial flagellar capping proteins adopt diverse oligomeric states.
Elife, 5, 2016
|
|
5I9Y
 
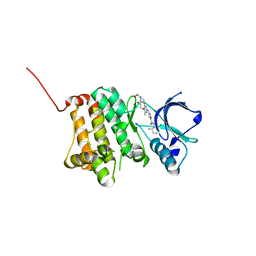 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with dasatinib | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.228 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
6GJ6
 
 | | CRYSTAL STRUCTURE OF KRAS G12D (GPPCP) IN COMPLEX WITH 18 | | Descriptor: | (3~{S})-3-[2-[(dimethylamino)methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Kessler, D, Mcconnell, D.M, Mantoulidis, A, Phan, J. | | Deposit date: | 2018-05-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Drugging an undruggable pocket on KRAS.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GJ8
 
 | | CRYSTAL STRUCTURE OF KRAS G12D (GPPCP) IN COMPLEX WITH BI 2852 | | Descriptor: | (3~{S})-3-[2-[[[1-[(1-methylimidazol-4-yl)methyl]indol-6-yl]methylamino]methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Kessler, D, Mcconnell, D.M, Mantoulidis, A. | | Deposit date: | 2018-05-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Drugging an undruggable pocket on KRAS.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5IO9
 
 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
5D24
 
 | | First bromodomain of BRD4 bound to inhibitor XD26 | | Descriptor: | 1,2-ETHANEDIOL, 4-acetyl-N-[3-(2-amino-2-oxoethoxy)phenyl]-3-ethyl-5-methyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Huegle, M, Gerhardt, S. | | Deposit date: | 2015-08-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
5D3H
 
 | | First bromodomain of BRD4 bound to inhibitor XD29 | | Descriptor: | Bromodomain-containing protein 4, N-[5-(diethylsulfamoyl)-2-hydroxyphenyl]-3-ethyl-4-(hydroxyacetyl)-5-methyl-1H-pyrrole-2-carboxamide | | Authors: | Wohlwend, D, Huegle, M, Weitzel, G. | | Deposit date: | 2015-08-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
8B7A
 
 | | Tubulin - maytansinoid - 4 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Boiarska, Z, Perez-Pena, H, Abel, A.-C, Marzullo, P, Alvarez-Bernad, B, Bonato, F, Santini, B, Horvath, D, Lucena-Agell, D, Vasile, F, Sironi, M, Diaz, F.J, Steinmetz, M.O, Prota, A.E, Piaraccini, S, Passarella, D. | | Deposit date: | 2022-09-29 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Maytansinol Functionalization: Towards Useful Probes for Studying Microtubule Dynamics.
Chemistry, 29, 2023
|
|
8B7C
 
 | | Tubulin-maytansinoid-12 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Boiarska, Z, Perez-Pena, H, Abel, A.-C, Marzullo, P, Alvarez-Bernad, B, Bonato, F, Santini, B, Horvath, D, Lucena-Agell, D, Vasile, F, Sironi, M, Diaz, J.F, Steinmetz, M.O, Prota, A.E, Pieraccini, S, Passarella, D. | | Deposit date: | 2022-09-29 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Maytansinol Functionalization: Towards Useful Probes for Studying Microtubule Dynamics.
Chemistry, 29, 2023
|
|
8B7B
 
 | | Tubulin - maytansinoid - 6 complex | | Descriptor: | (~{Z})-11-[[(1~{R},2~{R},3~{S},5~{S},6~{S},16~{E},18~{E},20~{R},21~{S})-11-chloranyl-12,20-dimethoxy-2,5,9,16-tetramethyl-21-oxidanyl-8,23-bis(oxidanylidene)-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1^{10,14}.0^{3,5}]hexacosa-10(26),11,13,16,18-pentaen-6-yl]oxy]-11-oxidanylidene-undec-2-enoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Boiarska, Z, Perez-Pena, H, Abel, A.-C, Marzullo, P, Alvarez-Bernad, B, Bonato, F, Santini, B, Horvath, D, Lucena-Agell, D, Vasile, F, Sironi, M, Diaz, J.F, Steinmetz, M.O, Prota, A.E, Pieraccini, S, Passarella, D. | | Deposit date: | 2022-09-29 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Maytansinol Functionalization: Towards Useful Probes for Studying Microtubule Dynamics.
Chemistry, 29, 2023
|
|
1D4L
 
 | | HIV-1 PROTEASE COMPLEXED WITH A MACROCYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | (10S,13S,1'R)-13-[1'-HYDROXY-2'-(N-P-AMINOBENZENESULFONYL-1''-AMINO-3''-METHYLBUTYL)ETHYL]-8,11-DIOXO-10-ISOPROPYL-2-OXA-9,12-DIAZABICYCLO [13.2.2]NONADECA-15,17,18-TRIENE, HIV-1 PROTEASE, SULFATE ION | | Authors: | Tyndall, J.D, Reid, R.C, Tyssen, D.P, Jardine, D.K, Todd, B, Passmore, M, March, D.R, Pattenden, L.K, Alewood, D, Hu, S.H, Alewood, P.F, Birch, C.J, Martin, J.L, Fairlie, D.P. | | Deposit date: | 1999-10-04 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Synthesis, stability, antiviral activity, and protease-bound structures of substrate-mimicking constrained macrocyclic inhibitors of HIV-1 protease.
J.Med.Chem., 43, 2000
|
|
5D7W
 
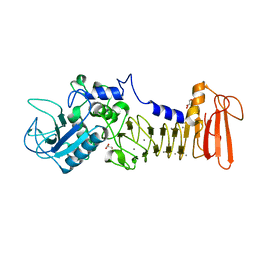 | | Crystal structure of serralysin | | Descriptor: | CALCIUM ION, GLYCEROL, Serralysin, ... | | Authors: | Wu, D, Ran, T, Xu, D.Q, Wang, W. | | Deposit date: | 2015-08-14 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of a thermostable serralysin from Serratia sp. FS14 at 1.1 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5ZB5
 
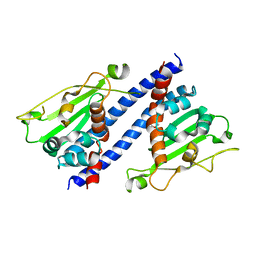 | | The structural basis of histone chaperoneVps75 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NAP family histone chaperone vps75 | | Authors: | Chen, Y, Zhang, Y, Dou, Y, Wang, M, Xu, S, Jiang, H, Limper, A, Su, D. | | Deposit date: | 2018-02-09 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural basis for the acetylation of histone H3K9 and H3K27 mediated by the histone chaperone Vps75 inPneumocystis carinii.
Signal Transduct Target Ther, 4, 2019
|
|
4ZK7
 
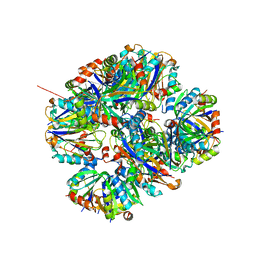 | | Crystal structure of rescued two-component self-assembling tetrahedral cage T33-31 | | Descriptor: | Chorismate mutase, Divalent-cation tolerance protein CutA | | Authors: | Liu, Y, Cascio, D, Sawaya, M.R, Bale, J, Collazo, M.J, Park, R, King, N, Baker, D, Yeates, T. | | Deposit date: | 2015-04-30 | | Release date: | 2015-07-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a designed tetrahedral protein assembly variant engineered to have improved soluble expression.
Protein Sci., 24, 2015
|
|
5CVI
 
 | | Structure of the manganese regulator SloR | | Descriptor: | 1,2-ETHANEDIOL, SloR, ZINC ION | | Authors: | Nye, D, Glasfeld, A. | | Deposit date: | 2015-07-27 | | Release date: | 2015-08-12 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | SloR structure and DNA binding studies inform the SloR-SRE interaction in Streptococcus mutans
to be published
|
|
5D3J
 
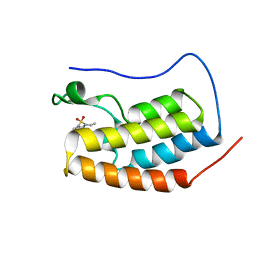 | | First bromodomain of BRD4 bound to inhibitor XD33 | | Descriptor: | 4-acetyl-N-[3-(diethylsulfamoyl)phenyl]-3-ethyl-5-methyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Huegle, M. | | Deposit date: | 2015-08-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
5D3R
 
 | | First bromodomain of BRD4 bound to inhibitor XD42 | | Descriptor: | 4-acetyl-N-[3-(azepan-1-ylsulfonyl)phenyl]-5-methyl-3-propyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Huegle, M. | | Deposit date: | 2015-08-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
2INX
 
 | | Crystal Structure of Ketosteroid Isomerase D40N from Pseudomonas putida (pKSI) with bound 2,6-difluorophenol | | Descriptor: | 2,6-DIFLUOROPHENOL, Steroid delta-isomerase | | Authors: | Martinez Caaveiro, J.M, Pybus, B, Ringe, D, Petsko, G.A, Sigala, P, Kraut, D, Herschlag, D. | | Deposit date: | 2006-10-09 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Testing geometrical discrimination within an enzyme active site: constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole.
J.Am.Chem.Soc., 130, 2008
|
|
6I7E
 
 | | Plasmodium falciparum Myosin A, Pre-powerstroke | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin-A, ... | | Authors: | Robert-Paganin, J, Auguin, D, Moussaoui, D, Jousset, G, Baum, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.492 Å) | | Cite: | Plasmodium myosin A drives parasite invasion by an atypical force generating mechanism.
Nat Commun, 10, 2019
|
|
