4DWO
 
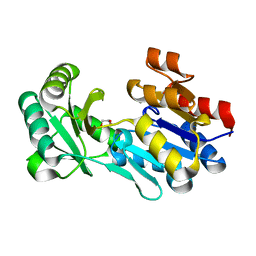 | | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Mg crystal form II | | Descriptor: | GLYCEROL, Haloacid dehalogenase-like hydrolase, MAGNESIUM ION | | Authors: | Vetting, M.W, Wasserman, S.R, Morisco, L.L, Sojitra, S, Allen, K.N, Dunaway-Mariano, D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-26 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Mg crystal form II
To be Published
|
|
3KHD
 
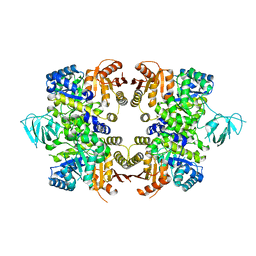 | | Crystal Structure of PFF1300w. | | Descriptor: | Pyruvate kinase | | Authors: | Wernimont, A.K, Hutchinson, A, Hassanali, A, Mackenzie, F, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Pizarro, J.C, Bakszt, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-30 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of PFF1300w.
To be Published
|
|
2OG9
 
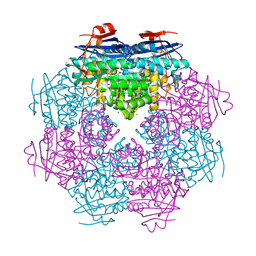 | |
2OHF
 
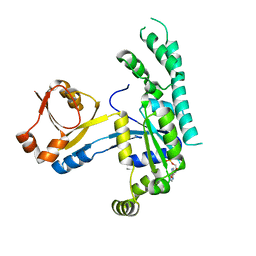 | |
2NWA
 
 | | X-ray Crystal Structure of Protein ytmB from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR466 | | Descriptor: | Hypothetical protein ytmB, SULFATE ION | | Authors: | Zhou, W, Forouhar, F, Seetharaman, J, Wang, D, Cunningham, K, Ma, L.-C, Fang, Y, Xiao, R, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2007-01-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Hypothetical protein YtmB from Bacillus subtilis subsp. (subtilis str. 168), Northeast Structural Genomics target SR466
To be Published
|
|
6SQ2
 
 | |
3KJS
 
 | | Crystal Structure of T. cruzi DHFR-TS with 3 high affinity DHFR inhibitors: DQ1 inhibitor complex | | Descriptor: | 1,2-ETHANEDIOL, Dihydrofolate reductase-thymidylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2009-11-03 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthesis and characterization of potent inhibitors of Trypanosoma cruzi dihydrofolate reductase.
Bioorg.Med.Chem., 18, 2010
|
|
3KKZ
 
 | | Crystal structure of the Q5LES9_BACFN protein from Bacteroides fragilis. Northeast Structural Genomics Consortium Target BfR250. | | Descriptor: | S-ADENOSYLMETHIONINE, uncharacterized protein Q5LES9 | | Authors: | Vorobiev, S, Neely, H, Seetharaman, J, Wang, D, Ciccosanti, C, Foote, E.L, Sahdev, S, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-06 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.677 Å) | | Cite: | Crystal structure of the Q5LES9_BACFN protein from Bacteroides fragilis.
To be Published
|
|
6SBS
 
 | | YtrA from Sulfolobus acidocaldarius, a GntR-family transcription factor | | Descriptor: | Regulatory protein | | Authors: | Lemmens, L, Valegard, K, Lindas, A.C, Peeters, E, Maes, D. | | Deposit date: | 2019-07-22 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | YtrASa, a GntR-Family Transcription Factor, Represses Two Genetic Loci Encoding Membrane Proteins inSulfolobus acidocaldarius.
Front Microbiol, 10, 2019
|
|
3KVE
 
 | | Structure of native L-amino acid oxidase from Vipera ammodytes ammodytes: stabilization of the quaternary structure by divalent ions and structural changes in the dynamic active site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid oxidase, ... | | Authors: | Gergiova, D, Murakami, M.T, Perbandt, M, Arni, R.K, Betzel, C. | | Deposit date: | 2009-11-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of native L-amino acid oxidase from Vipera ammodytes ammodytes: stabilization of the quaternary structure by divalent ions and structural changes in the dynamic active site
To be Published
|
|
8Q4G
 
 | | Thin filament from FIB milled relaxed left ventricular mouse myofibrils | | Descriptor: | Actin, alpha cardiac muscle 1, Tropomyosin alpha-1 chain | | Authors: | Tamborrini, D, Wang, Z, Wagner, T, Tacke, S, Stabrin, M, Grange, M, Kho, A.L, Bennet, P, Rees, M, Gautel, M, Raunser, S. | | Deposit date: | 2023-08-06 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure of the native myosin filament in the relaxed cardiac sarcomere.
Nature, 623, 2023
|
|
8Q3C
 
 | | Structure of Selenomonas ruminantium lactate dehydrogenase I85R mutant | | Descriptor: | CHLORIDE ION, L-lactate dehydrogenase, NITRATE ION, ... | | Authors: | Bertrand, Q, Coquille, S, Iorio, A, Sterpone, F, Madern, D. | | Deposit date: | 2023-08-03 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical, structural and dynamical characterizations of the lactate dehydrogenase from Selenomonas ruminantium provide information about an intermediate evolutionary step prior to complete allosteric regulation acquisition in the super family of lactate and malate dehydrogenases.
J.Struct.Biol., 215, 2023
|
|
2OIZ
 
 | |
2NY5
 
 | | HIV-1 gp120 Envelope Glycoprotein (M95W, W96C, I109C, T257S, V275C, S334A, S375W, Q428C, A433M) Complexed with CD4 and Antibody 17b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Zhou, T, Xu, L, Dey, B, Hessell, A.J, Van Ryk, D, Xiang, S.H, Yang, X, Zhang, M.Y, Zwick, M.B, Arthos, J, Burton, D.R, Dimitrov, D.S, Sodroski, J, Wyatt, R, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural definition of a conserved neutralization epitope on HIV-1 gp120.
Nature, 445, 2007
|
|
6SRW
 
 | | Kemp Eliminase HG3.17 mutant Q50F, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, GLYCEROL, Kemp Eliminase HG3.17 Q50F | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
6SS4
 
 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0021181 | | Descriptor: | Arginase-2, mitochondrial, Fab C0021181 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
2NUU
 
 | | Regulating the Escherichia coli ammonia channel: the crystal structure of the AmtB-GlnK complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ammonia channel, Nitrogen regulatory protein P-II 2 | | Authors: | Conroy, M.J, Durand, A, Lupo, D, Li, X.-D, Bullough, P.A, Winkler, F.K, Merrick, M. | | Deposit date: | 2006-11-09 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6SDR
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Oxidized form | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Formate dehydrogenase, alpha subunit, ... | | Authors: | Oliveira, A.R, Mota, C, Mourato, C, Domingos, R.M, Santos, M.F.A, Gesto, D, Guigliarelli, B, Santos-Silva, T, Romao, M.J, Pereira, I.C. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards the mechanistic understanding of enzymatic CO2 reduction
Acs Catalysis, 2020
|
|
6SE5
 
 | |
3KP1
 
 | | Crystal structure of ornithine 4,5 aminomutase (Resting State) | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-ornithine aminomutase E component, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
2O2K
 
 | | Crystal Structure of the Activation Domain of Human Methionine Synthase Isoform/Mutant D963E/K1071N | | Descriptor: | Methionine synthase | | Authors: | Wolthers, K.R, Toogood, H.S, Jowitt, T.A, Marshall, K.R, Leys, D, Scrutton, N.S. | | Deposit date: | 2006-11-30 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and solution characterization of the activation domain of human methionine synthase
Febs J., 274, 2007
|
|
2NXZ
 
 | | HIV-1 gp120 Envelope Glycoprotein (T257S, S334A, S375W) Complexed with CD4 and Antibody 17b | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, ... | | Authors: | Zhou, T, Xu, L, Dey, B, Hessell, A.J, Van Ryk, D, Xiang, S.H, Yang, X, Zhang, M.Y, Zwick, M.B, Arthos, J, Burton, D.R, Dimitrov, D.S, Sodroski, J, Wyatt, R, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural definition of a conserved neutralization epitope on HIV-1 gp120.
Nature, 445, 2007
|
|
6SI6
 
 | | N-terminal domain of Drosophila X virus VP3 | | Descriptor: | GLYCEROL, IMIDAZOLE, Structural polyprotein | | Authors: | Ferrero, D.S, Garriga, D, Guerra, P, Uson, I, Verdaguer, N. | | Deposit date: | 2019-08-08 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and dsRNA-binding activity of the Birnavirus Drosophila X Virus VP3 protein.
J.Virol., 2020
|
|
3KRU
 
 | | Crystal Structure of the Thermostable Old Yellow Enzyme from Thermoanaerobacter pseudethanolicus E39 | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, NADH:flavin oxidoreductase/NADH oxidase | | Authors: | Adalbjornsson, B.V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2009-11-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biocatalysis with thermostable enzymes: structure and properties of a thermophilic 'ene'-reductase related to old yellow enzyme.
Chembiochem, 11, 2010
|
|
3KOW
 
 | | Crystal Structure of ornithine 4,5 aminomutase backsoaked complex | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-ornithine aminomutase E component, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
