8QG1
 
 | |
3VTX
 
 | | Crystal structure of MamA protein | | Descriptor: | GLYCEROL, MamA | | Authors: | Zeytuni, N, Baran, D, Davidov, G, Zarivach, R. | | Deposit date: | 2012-06-08 | | Release date: | 2012-10-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inter-phylum structural conservation of the magnetosome-associated TPR-containing protein, MamA
J.Struct.Biol., 180, 2012
|
|
1A6I
 
 | | TET REPRESSOR, CLASS D VARIANT | | Descriptor: | TETRACYCLINE REPRESSOR PROTEIN CLASS D | | Authors: | Orth, P, Cordes, F, Schnappinger, D, Hillen, W, Saenger, W, Hinrichs, W. | | Deposit date: | 1998-02-25 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational changes of the Tet repressor induced by tetracycline trapping.
J.Mol.Biol., 279, 1998
|
|
7VLT
 
 | | Structure of SUR2B in complex with Mg-ATP/ADP and levcromakalim | | Descriptor: | (3S,4R)-2,2-dimethyl-3-oxidanyl-4-(2-oxidanylidenepyrrolidin-1-yl)-3,4-dihydrochromene-6-carbonitrile, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
5LI7
 
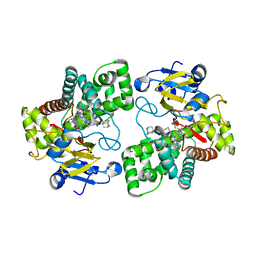 | | Crystal structure of Mycobacterium tuberculosis CYP126A1 in complex with 1-(3-(1H-imidazol-1-yl)propyl)-3-((3s,5s,7s)-adamantan-1-yl)urea | | Descriptor: | 1-(1-adamantyl)-3-(3-imidazol-1-ylpropyl)urea, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 126 | | Authors: | Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2016-07-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Characterization and Ligand/Inhibitor Identification Provide Functional Insights into the Mycobacterium tuberculosis Cytochrome P450 CYP126A1.
J. Biol. Chem., 292, 2017
|
|
7VLR
 
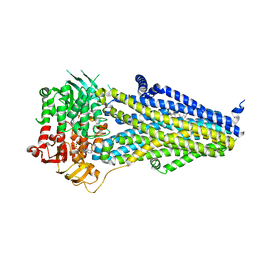 | | Structure of SUR2B in complex with Mg-ATP/ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
3W0S
 
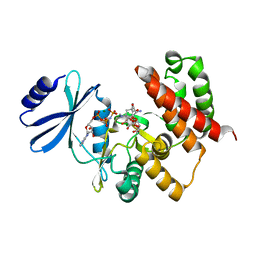 | | Crystal structure of aminoglycoside phosphotransferase APH(4)-Ia, ternary complex with AMP-PNP and hygromycin B | | Descriptor: | HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase, MAGNESIUM ION, ... | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
8QH7
 
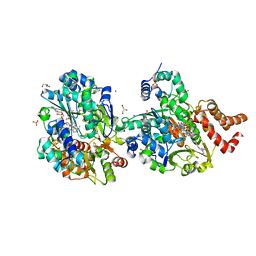 | | Crystal structure of respiratory Complex I subunits NuoEF from Aquifex aeolicus bound to reduced 3-acetylpyridine adenine dinucleotide (without reducing agent) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYL PYRIDINE ADENINE DINUCLEOTIDE, ... | | Authors: | Wohlwend, D, Friedrich, T. | | Deposit date: | 2023-09-06 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of 3-acetylpyridine adenine dinucleotide and ADP-ribose bound to the electron input module of respiratory complex I.
Structure, 32, 2024
|
|
6ARD
 
 | |
6ARK
 
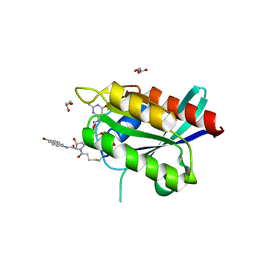 | | Crystal Structure of compound 10 covalently bound to K-Ras G12C | | Descriptor: | (3R)-N-(6-bromonaphthalen-2-yl)-3-hydroxy-1-propanoyl-L-prolinamide, GLYCEROL, GTPase KRas, ... | | Authors: | Nnadi, C.I, Jenkins, M.L, Gentile, D.R, Bateman, L.A, Zaidman, D, Balius, T.E, Nomura, D.K, Burke, J.E, Shokat, K.M, London, N. | | Deposit date: | 2017-08-22 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel K-Ras G12C Switch-II Covalent Binders Destabilize Ras and Accelerate Nucleotide Exchange.
J Chem Inf Model, 58, 2018
|
|
8QHK
 
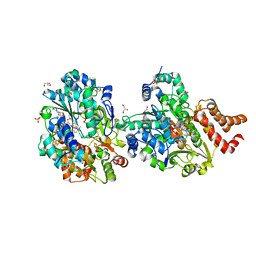 | | Crystal structure of reduced respiratory Complex I subunits NuoEF from Aquifex aeolicus bound to reduced 3-acetylpyridine adenine dinucleotide | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ACETYL PYRIDINE ADENINE DINUCLEOTIDE, REDUCED, ... | | Authors: | Wohlwend, D, Friedrich, T, Bucka, S. | | Deposit date: | 2023-09-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of 3-acetylpyridine adenine dinucleotide and ADP-ribose bound to the electron input module of respiratory complex I.
Structure, 32, 2024
|
|
7VZB
 
 | | Cryo-EM structure of C22:0-CoA bound human very long-chain fatty acid ABC transporter ABCD1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Peroxisomal Membrane Protein related,ATP-binding cassette sub-family D member 1, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] docosanethioate | | Authors: | Chen, Z.P, Xu, D, Wang, L, Mao, Y.X, Yang, L, Cheng, M.T, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2021-11-15 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural basis of substrate recognition and translocation by human very long-chain fatty acid transporter ABCD1.
Nat Commun, 13, 2022
|
|
8QCX
 
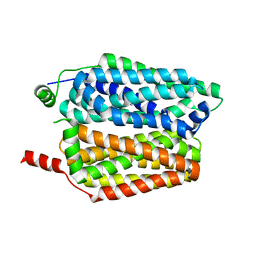 | |
7VLU
 
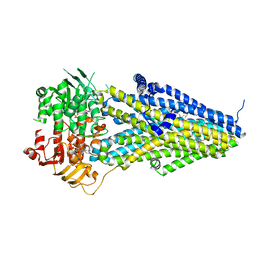 | | Structure of SUR2A in complex with Mg-ATP/ADP and P1075 | | Descriptor: | 1-cyano-2-(2-methylbutan-2-yl)-3-pyridin-3-yl-guanidine, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
8QGW
 
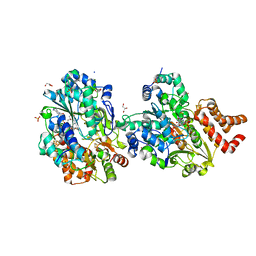 | |
7VLS
 
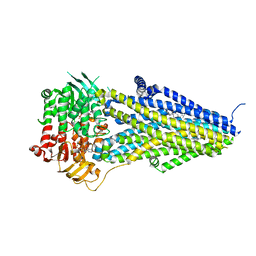 | | Structure of SUR2B in complex with MgATP/ADP and P1075 | | Descriptor: | 1-cyano-2-(2-methylbutan-2-yl)-3-pyridin-3-yl-guanidine, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
7V7W
 
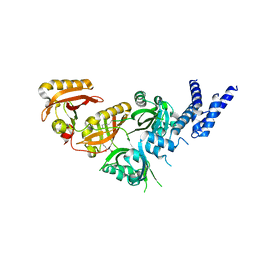 | | Crystal Structure of the Heterodimeric HIF-3a:ARNT Complex with oleoylethanolamide (OEA) | | Descriptor: | (Z)-N-(2-hydroxyethyl)octadec-9-enamide, Aryl hydrocarbon receptor nuclear translocator, Hypoxia-inducible factor 3-alpha | | Authors: | Diao, X, Ren, X, Li, F.W, Zhang, M, Sun, X, Wu, D. | | Deposit date: | 2021-08-21 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Nat Commun, 13, 2022
|
|
7VX8
 
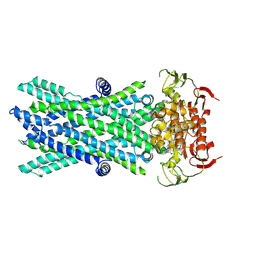 | | Cryo-EM structure of ATP-bound human very long-chain fatty acid ABC transporter ABCD1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Peroxisomal Membrane Protein related,ATP-binding cassette sub-family D member 1 | | Authors: | Chen, Z.P, Xu, D, Wang, L, Mao, Y.X, Yang, L, Cheng, M.T, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2021-11-12 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of substrate recognition and translocation by human very long-chain fatty acid transporter ABCD1.
Nat Commun, 13, 2022
|
|
7VWC
 
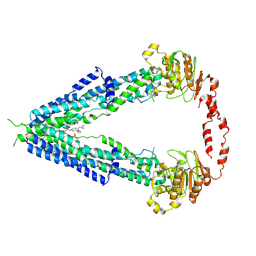 | | Cryo-EM structure of human very long-chain fatty acid ABC transporter ABCD1 | | Descriptor: | Peroxisomal Membrane Protein related,ATP-binding cassette sub-family D member 1, [(2R)-3-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-2-oxidanyl-propyl] octadecanoate | | Authors: | Chen, Z.P, Xu, D, Wang, L, Mao, Y.X, Yang, L, Cheng, M.T, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2021-11-10 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis of substrate recognition and translocation by human very long-chain fatty acid transporter ABCD1.
Nat Commun, 13, 2022
|
|
3NW3
 
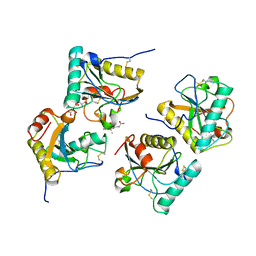 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with the PGN Fragment at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, ALANINE, D-GLUTAMINE, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-07-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multiligand specificity of pathogen-associated molecular pattern-binding site in peptidoglycan recognition protein
J.Biol.Chem., 286, 2011
|
|
3NWE
 
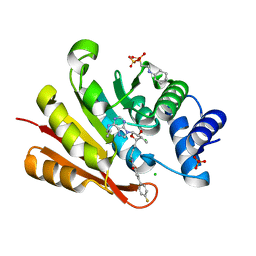 | | Rat COMT in complex with a methylated desoxyribose bisubstrate-containing inhibitor avoids hydroxyl group | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 5-(4-fluorophenyl)-2,3-dihydroxy-N-[(E)-3-[(2R,3R,4R,5R)-4-hydroxy-3-methyl-5-[6-(propylamino)purin-9-yl]oxolan-2-yl]prop-2-enyl]benzamide, CHLORIDE ION, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2010-07-09 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catechol-O-methyltransferase in complex with substituted 3'-deoxyribose bisubstrate inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6B3J
 
 | | 3.3 angstrom phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex | | Descriptor: | Exendin-P5, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liang, Y.L, Khoshouei, M, Glukhova, A, Furness, S.G.B, Koole, C, Zhao, P, Clydesdale, L, Thal, D.M, Radjainia, M, Danev, R, Baumeister, W, Wang, M.W, Miller, L.J, Christopoulos, A, Sexton, P.M, Wootten, D. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-21 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex.
Nature, 555, 2018
|
|
8QCY
 
 | |
6ARU
 
 | | Structure of Cetuximab Fab mutant in complex with EGFR extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab mutant heavy chain Fab fragment,Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Christie, M, Christ, D. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Cetuximab Fab mutant in complex with EGFR extracellular domain
To Be Published
|
|
3W0P
 
 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia (D198A), ternary complex with ADP and hygromycin B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
