5MR3
 
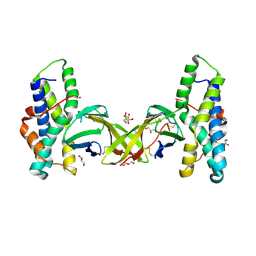 | | Crystal structure of red abalone egg VERL repeat 2 with linker in complex with sperm lysin at 1.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Egg-lysin, ... | | Authors: | Nishimura, K, Raj, I, Sadat Al-Hosseini, H, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
2A55
 
 | | Solution structure of the two N-terminal CCP modules of C4b-binding protein (C4BP) alpha-chain. | | Descriptor: | C4b-binding protein | | Authors: | Jenkins, H.T, Mark, L, Ball, G, Lindahl, G, Uhrin, D, Blom, A.M, Barlow, P.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-12-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Human C4b-binding Protein, Structural Basis for Interaction with Streptococcal M Protein, a Major Bacterial Virulence Factor
J.Biol.Chem., 281, 2006
|
|
2XJP
 
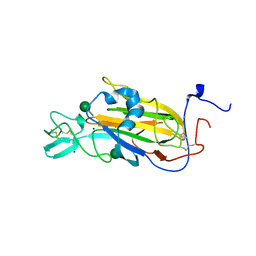 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae in complex with calcium and mannose | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5MN6
 
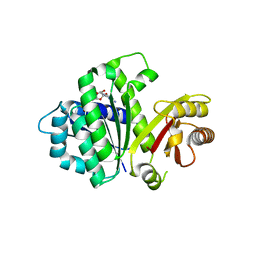 | | S. aureus FtsZ 12-316 F138A GDP Closed form (3FCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
2A77
 
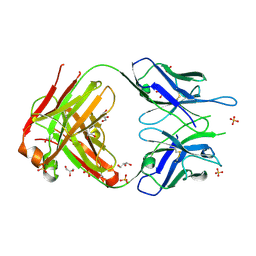 | | Anti-Cocaine Antibody 7.5.21, Crystal Form II | | Descriptor: | GLYCEROL, Immunoglobulin Heavy Chain, Immunoglobulin Light Chain, ... | | Authors: | Pozharski, E, Hewagama, A, Shanafelt, A, Ringe, D, Petsko, G.A. | | Deposit date: | 2005-07-04 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Flexibility of Packing: Four Crystal Forms of an Anti-Cocaine Antibody 7.5.21
To be Published
|
|
6UKP
 
 | | Apo mBcs1 | | Descriptor: | Mitochondrial chaperone BCS1 | | Authors: | Tang, W.K, Borgnia, M.J, Hsu, A.L, Xia, D. | | Deposit date: | 2019-10-05 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structures of AAA protein translocase Bcs1 suggest translocation mechanism of a folded protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2XF5
 
 | | Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. | | Descriptor: | GP23.1 | | Authors: | Veesler, D, Blangy, S, Lichiere, J, Ortiz-Lombardia, M, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-11 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp23.1, A Putative Chaperone.
Protein Sci., 19, 2010
|
|
2XGB
 
 | | Crystal structure of Barley Beta-Amylase complexed with 2,3- epoxypropyl-alpha-D-glucopyranoside | | Descriptor: | (2R)-oxiran-2-ylmethyl alpha-D-glucopyranoside, 1,2-ETHANEDIOL, BETA-AMYLASE | | Authors: | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | Deposit date: | 2010-06-02 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Chemical Genetics and Cereal Starch Metabolism: Structural Basis of the Non-Covalent and Covalent Inhibition of Barley Beta-Amylase.
Mol.Biosyst., 7, 2011
|
|
2XJS
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae in complex with calcium and a1,2-mannobiose | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6UCY
 
 | | Multi-conformer model of Ketosteroid Isomerase from Pseudomonas Putida (pKSI) bound to 4-Androstenedione at 250 K | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Yabukarski, F, Herschlag, D, Biel, J.T, Fraser, J.S. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray-derived conformational ensembles.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2XC8
 
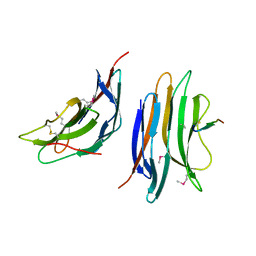 | | Crystal structure of the gene 22 product of the Bacillus subtilis SPP1 phage | | Descriptor: | GENE 22 PRODUCT | | Authors: | Veesler, D, Blangy, S, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-04-19 | | Release date: | 2010-06-09 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp22 Shares Fold Similarity with a Domain of Lactococcal Phage P2 Rbp.
Protein Sci., 19, 2010
|
|
6UD9
 
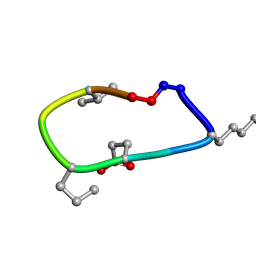 | | S2 symmetric peptide design number 2, Morticia | | Descriptor: | S2-2, Morticia | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
2XQK
 
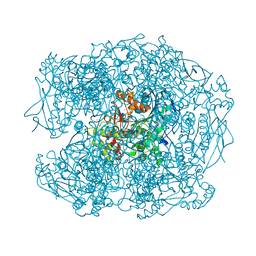 | | X-ray Structure of human butyrylcholinesterase inhibited by pure enantiomer VX-(S) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wandhammer, M, Carletti, E, Gillon, E, Masson, P, Goeldner, M, Noort, D, Nachon, F. | | Deposit date: | 2010-09-02 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Study of the Complex Stereoselectivity of Human Butyrylcholinesterase for the Neurotoxic V-Agents.
J.Biol.Chem., 286, 2011
|
|
5MUN
 
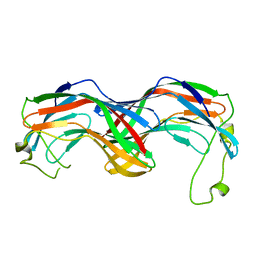 | | Structural insight into zymogenic latency of gingipain K from Porphyromonas gingivalis. | | Descriptor: | AZIDE ION, Lys-gingipain W83 | | Authors: | Pomowski, A, Uson, I, Nowakovska, Z, Veillard, F, Sztukowska, M.N, Guevara, T, Goulas, T, Mizgalska, D, Nowak, M, Potempa, B, Huntington, J.A, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights unravel the zymogenic mechanism of the virulence factor gingipain K from Porphyromonas gingivalis, a causative agent of gum disease from the human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
2XHD
 
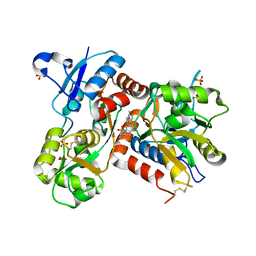 | | Crystal structure of N-((2S)-5-(6-fluoro-3-pyridinyl)-2,3-dihydro-1H- inden-2-yl)-2-propanesulfonamide in complex with the ligand binding domain of the human GluA2 receptor | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N-[(2S)-5-(6-FLUORO-3-PYRIDINYL)-2,3-DIHYDRO-1H-INDEN-2-YL]-2-PROPANESULFONAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Andreotti, D, Ballantine, S, Bax, B.D, Harris, A.J, Harker, A.J, Lund, J, Melarange, R, Mingardi, A, Mookherjee, C, Mosley, J, Neve, M, Oliosi, B, Profeta, R, Smith, K.J, Smith, P.W, Spada, S, Thewlis, K.M, Yusaf, S.P. | | Deposit date: | 2010-06-14 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of N-[(2S)-5-(6-Fluoro-3-Pyridinyl)-2,3-Dihydro-1H-Inden-2-Yl]-2-Propanesulfonamide, a Novel Clinical Ampa Receptor Positive Modulator.
J.Med.Chem., 53, 2010
|
|
8AOL
 
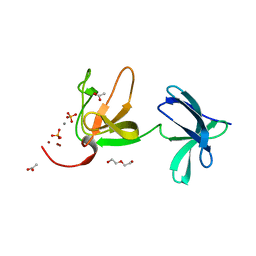 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain III (aa 363-499) | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sagmeister, T, Damisch, E, Eder, M, Dordic, A, Vejzovic, D, Pavkov-Keller, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6UCC
 
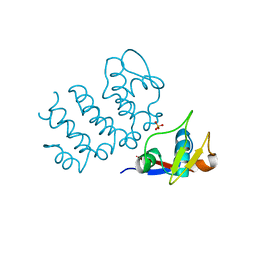 | | Structure of human PACRG-MEIG1 complex (limited proteolysis) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Meiosis expressed gene 1 protein homolog, PHOSPHATE ION, ... | | Authors: | Khan, N, Croteau, N, Pelletier, D, Veyron, S, Trempe, J.F. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human PACRG in complex with MEIG1
Biorxiv, 2019
|
|
2A9H
 
 | | NMR structural studies of a potassium channel / charybdotoxin complex | | Descriptor: | Voltage-gated potassium channel, charybdotoxin | | Authors: | Yu, L, Sun, C, Song, D, Shen, J, Xu, N, Gunasekera, A, Hajduk, P.J, Olejniczak, E.T. | | Deposit date: | 2005-07-11 | | Release date: | 2006-01-10 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies of a potassium channel-charybdotoxin complex.
Biochemistry, 44, 2005
|
|
2XEW
 
 | | Crystal structure of K11-linked diubiquitin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Bremm, A, Freund, S.M.V, Komander, D. | | Deposit date: | 2010-05-18 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lys11-Linked Ubiquitin Chains Adopt Compact Conformations and are Preferentially Hydrolyzed by the Deubiquitinase Cezanne
Nat.Struct.Mol.Biol., 17, 2010
|
|
6UCQ
 
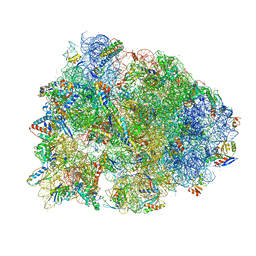 | | Crystal structure of the Thermus thermophilus 70S ribosome recycling complex | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhou, D, Tanzawa, T, Gagnon, M.G, Lin, J. | | Deposit date: | 2019-09-17 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for ribosome recycling by RRF and tRNA.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2XTS
 
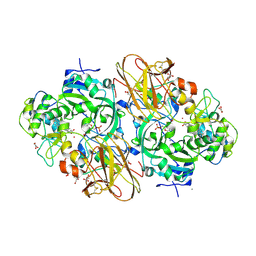 | | Crystal Structure of the Sulfane Dehydrogenase SoxCD from Paracoccus pantotrophus | | Descriptor: | CALCIUM ION, COBALT (II) ION, CYTOCHROME, ... | | Authors: | Zander, U, Faust, A, Klink, B.U, de Sanctis, D, Panjikar, S, Quentmeier, A, Bardischewski, F, Friedrich, C.G, Scheidig, A.J. | | Deposit date: | 2010-10-12 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural Basis for the Oxidation of Protein-Bound Sulfur by the Sulfur Cycle Molybdohemo-Enzyme Sulfane Dehydrogenase Soxcd.
J.Biol.Chem., 286, 2011
|
|
1ZZP
 
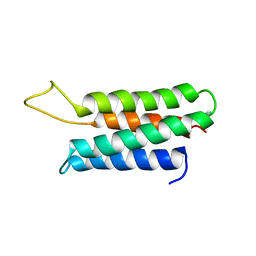 | | Solution structure of the F-actin binding domain of Bcr-Abl/c-Abl | | Descriptor: | Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Hantschel, O, Wiesner, S, Guttler, T, Mackereth, C.D, Rix, L.L.R, Mikes, Z, Dehne, J, Gorlich, D, Sattler, M, Superti-Furga, G. | | Deposit date: | 2005-06-14 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Cytoskeletal Association of Bcr-Abl/c-Abl.
Mol.Cell, 19, 2005
|
|
5MWO
 
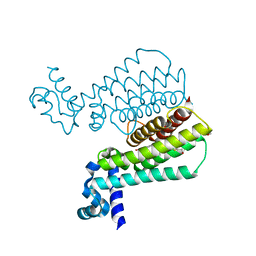 | | Structure of Mycobacterium Tuberculosis Transcriptional Regulatory Repressor Protein (EthR) in complex with fragment 7E8. | | Descriptor: | (5-methyl-1-benzothiophen-2-yl)methanol, 1,2-ETHANEDIOL, HTH-type transcriptional regulator EthR | | Authors: | Mendes, V, Chan, D.S.-H, Thomas, S.E, McConnell, B, Matak-Vinkovic, D, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2017-01-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Fragment Screening against the EthR-DNA Interaction by Native Mass Spectrometry.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2XP6
 
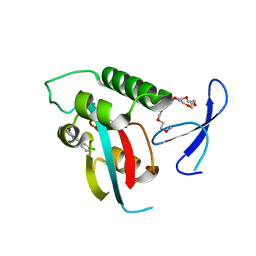 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 2-(3-CHLORO-PHENYL)-5-METHYL-1H-IMIDAZOLE-4-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6UPZ
 
 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 3 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
