6ZD2
 
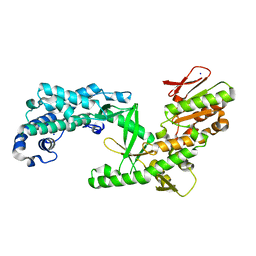 | | Structure of apo telomerase from Candida Tropicalis truncated from C-terminal domain | | Descriptor: | SODIUM ION, Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-13 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
6ZDQ
 
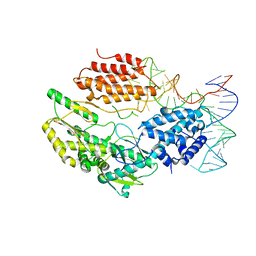 | | Structure of telomerase from Candida albicans in complexe with TWJ fragment of telomeric RNA | | Descriptor: | RNA, Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-15 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
6ZDU
 
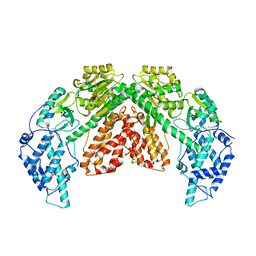 | | Structure of telomerase from Candida albicans in complexe with TWJ fragment of telomeric RNA | | Descriptor: | Chains: C,D, Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-15 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
7ORE
 
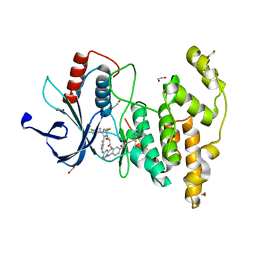 | | Crystal structure of JNK3 in complex with light-activated covalent inhibitor MR-II-249 with both non-covalent and covalent binding modes (compound 4) | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-N-[(5Z)-9-[[4-[5-(4-fluorophenyl)-3-methyl-2-methylsulfanyl-imidazol-4-yl]pyridin-2-yl]amino]-11,12-dihydrobenzo[c][1,2]benzodiazocin-2-yl]butanamide, Mitogen-activated protein kinase 10 | | Authors: | Chaikuad, A, Reynders, M, Trauner, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-05 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Controlling the Covalent Reactivity of a Kinase Inhibitor with Light.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6ZD1
 
 | | Structure of apo telomerase from Candida Tropicalis | | Descriptor: | Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-13 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
6HT0
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with compound 94 | | Descriptor: | 1,2-ETHANEDIOL, 1-cyclopropyl-~{N}-[2-[[(2~{S})-2-methylpyrrolidin-1-yl]methyl]-3~{H}-benzimidazol-5-yl]indazole-5-carboxamide, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an MLLT1/3 YEATS Domain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6TMY
 
 | | Crystal structure of isoform CBd of the basic phospholipase A2 subunit of crotoxin from Crotalus durissus terrificus | | Descriptor: | CHLORIDE ION, Phospholipase A2 crotoxin basic subunit CBc, SODIUM ION, ... | | Authors: | Nemecz, D, Ostrowski, M, Saul, F.A, Faure, G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Isoform CBd of the Basic Phospholipase A 2 Subunit of Crotoxin: Description of the Structural Framework of CB for Interaction with Protein Targets.
Molecules, 25, 2020
|
|
6ZD6
 
 | | Structure of apo telomerase from Candida Tropicalis | | Descriptor: | Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-13 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
6ZDP
 
 | | Structure of telomerase from Candida Tropicalis in complexe with TWJ fragment of telomeric RNA | | Descriptor: | Chains: B, POTASSIUM ION, Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-15 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
6YX8
 
 | | The structure of allophycocyanin from cyanobacterium Nostoc sp. WR13, the C2221 crystal form. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Patel, H.M, Roszak, A.W, Madamwar, D, Cogdell, R.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | The high resolution structure of allophycocyanin from cyanobacterium Nostoc sp. WR13
To Be Published
|
|
7P8K
 
 | | Crystal structure of in planta processed AvrRps4 in complex with the WRKY domain of RRS1 | | Descriptor: | Avirulence protein,Avirulence protein, Disease resistance protein RRS1, ZINC ION | | Authors: | Mukhi, N, Brown, H, Gorenkin, D, Ding, P, Bentham, A.R, Jones, J.D.G, Banfield, M.J. | | Deposit date: | 2021-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Perception of structurally distinct effectors by the integrated WRKY domain of a plant immune receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6YX7
 
 | | The high resolution structure of allophycocyanin from cyanobacterium Nostoc sp. WR13, the P21212 crystal form. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Patel, H.M, Roszak, A.W, Madamwar, D, Cogdell, R.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | The high resolution structure of allophycocyanin from cyanobacterium Nostoc sp. WR13
To Be Published
|
|
6HR4
 
 | |
7OSD
 
 | |
7P5M
 
 | | Cryo-EM structure of human TTYH2 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 2 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7OLZ
 
 | | Crystal structure of the SARS-CoV-2 RBD with neutralizing-VHHs Re5D06 and Re9F06 | | Descriptor: | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, Nanobody Re5D06, Nanobody Re9F06, ... | | Authors: | Aksu, M, Guttler, T, Gorlich, D. | | Deposit date: | 2021-05-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies.
Embo J., 40, 2021
|
|
6TL7
 
 | |
7P54
 
 | | Cryo-EM structure of human TTYH2 in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 2 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7P5J
 
 | | Cryo-EM structure of human TTYH1 in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 1 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
6TSR
 
 | | Marasmius oreades agglutinin (MOA) activated by manganese (II) and calcium | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin, CALCIUM ION, ... | | Authors: | Cordara, G, Manna, D, Krengel, U. | | Deposit date: | 2019-12-21 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of MOA in complex with a peptide fragment: A protease caught in flagranti .
Curr Res Struct Biol, 2, 2020
|
|
7ON5
 
 | | Crystal structure of the SARS-CoV-2 neutralizing nanobody Re5D06 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Nanobody Re5D06 | | Authors: | Aksu, M, Guttler, T, Gorlich, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies.
Embo J., 40, 2021
|
|
7OS8
 
 | | NMR SOLUTION STRUCTURE OF [Pro3,DLeu9]TL | | Descriptor: | PHE-VAL-PRO-TRP-PHE-SER-LYS-PHE-DLE-GLY-ARG-ILE-LEU-NH2 | | Authors: | Brancaccio, D, Carotenuto, A. | | Deposit date: | 2021-06-08 | | Release date: | 2021-08-11 | | Last modified: | 2021-08-25 | | Method: | SOLUTION NMR | | Cite: | First-in-Class Cyclic Temporin L Analogue: Design, Synthesis, and Antimicrobial Assessment.
J.Med.Chem., 64, 2021
|
|
6HYN
 
 | | Structure of ATG13 LIR motif bound to GABARAP | | Descriptor: | Autophagy-related protein 13,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
6DNK
 
 | | Human Stimulator of Interferon Genes | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Fernandez, D, Li, L, Ergun, S.L. | | Deposit date: | 2018-06-06 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition.
Cell, 178, 2019
|
|
6TTX
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 5 (ASI_M3M_051) | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)-~{N}-(piperidin-4-ylmethyl)oxolane-2-carboxamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
