1WRT
 
 | | NMR STUDY OF APO TRP REPRESSOR | | Descriptor: | APO TRP REPRESSOR | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structures of the Escherichia coli trp holo- and aporepressor.
J.Mol.Biol., 229, 1993
|
|
8DM3
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with Fab 4A8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 4A8 heavy chain, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
1WRS
 
 | | NMR STUDY OF HOLO TRP REPRESSOR | | Descriptor: | HOLO TRP REPRESSOR, TRYPTOPHAN | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structures of the Escherichia coli trp holo- and aporepressor.
J.Mol.Biol., 229, 1993
|
|
8AFB
 
 | | CRYSTAL STRUCTURE OF KRAS-G12C IN COMPLEX WITH COMPOUND 23 (BI-0474) | | Descriptor: | (4~{S})-2-azanyl-4-[3-[6-[(2~{S})-2,4-dimethylpiperazin-1-yl]-4-(4-prop-2-enoylpiperazin-1-yl)pyridin-2-yl]-1,2,4-oxadiazol-5-yl]-4-methyl-6,7-dihydro-5~{H}-1-benzothiophene-3-carbonitrile, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Boettcher, J, Kessler, D. | | Deposit date: | 2022-07-16 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Fragment Optimization of Reversible Binding to the Switch II Pocket on KRAS Leads to a Potent, In Vivo Active KRAS G12C Inhibitor.
J.Med.Chem., 65, 2022
|
|
8DM2
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein (focused refinement of NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
6X2L
 
 | | hEAAT3-IFS-Na | | Descriptor: | Excitatory amino acid transporter 3 | | Authors: | Qiu, B, Matthies, D, Boudker, O. | | Deposit date: | 2020-05-20 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport.
Sci Adv, 7, 2021
|
|
8AFD
 
 | | CRYSTAL STRUCTURE OF BIT-BLOCKED KRAS-G12V-S39C IN COMPLEX WITH COMPOUND 20a | | Descriptor: | (4~{S})-4-[3-(4-aminophenyl)-1,2,4-oxadiazol-5-yl]-2-azanyl-4-methyl-6,7-dihydro-5~{H}-1-benzothiophene-3-carbonitrile, 1H-benzimidazol-2-ylmethanethiol, GTPase KRas, ... | | Authors: | Boettcher, J, Kessler, D. | | Deposit date: | 2022-07-16 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.633 Å) | | Cite: | Fragment Optimization of Reversible Binding to the Switch II Pocket on KRAS Leads to a Potent, In Vivo Active KRAS G12C Inhibitor.
J.Med.Chem., 65, 2022
|
|
8AXP
 
 | |
6X2Z
 
 | | hEAAT3-OFS-Asp | | Descriptor: | ASPARTIC ACID, Excitatory amino acid transporter 3, SODIUM ION | | Authors: | Qiu, B, Matthies, D, Boudker, O. | | Deposit date: | 2020-05-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport.
Sci Adv, 7, 2021
|
|
6WQU
 
 | | CSL (RBPJ) bound to Notch3 RAM and DNA | | Descriptor: | DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*CP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), Neurogenic locus notch homolog protein 3, ... | | Authors: | Kovall, R.A, Gagliani, E, Hall, D. | | Deposit date: | 2020-04-29 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | PIM-induced phosphorylation of Notch3 promotes breast cancer tumorigenicity in a CSL-independent fashion.
J.Biol.Chem., 296, 2021
|
|
6X3E
 
 | | hEAAT3-Asymmetric-1o2i | | Descriptor: | ASPARTIC ACID, Excitatory amino acid transporter 3, SODIUM ION | | Authors: | Qiu, B, Matthies, D, Boudker, O. | | Deposit date: | 2020-05-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport.
Sci Adv, 7, 2021
|
|
8DM1
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
6X3F
 
 | | hEAAT3-IFS-Apo | | Descriptor: | CHOLINE ION, Excitatory amino acid transporter 3 | | Authors: | Qiu, B, Matthies, D, Boudker, O. | | Deposit date: | 2020-05-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport.
Sci Adv, 7, 2021
|
|
8DM4
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with Fab 4A8 (focused refinement of NTD and 4A8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 4A8 heavy chain, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
6WNM
 
 | | The structure of Pf4r from a superinfective isolate of the filamentous phage Pf4 of Pseudomonas aeruginosa PA01 | | Descriptor: | Pf4r, [(2R)-3-{[2-(carboxymethoxy)benzene-1-carbonyl]amino}-2-methoxypropyl](hydroxy)mercury | | Authors: | Michie, K.A, Norrian, P, Duggin, I.G, McDougald, D, Rice, S.A. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The Repressor C Protein, Pf4r, Controls Superinfection of Pseudomonas aeruginosa PAO1 by the Pf4 Filamentous Phage and Regulates Host Gene Expression.
Viruses, 13, 2021
|
|
7P84
 
 | | Crystal structure of L147A/I351A variant of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii in complex with ONB-SAM (2-nitro benzyme S-adenosyl-methionine) | | Descriptor: | MAGNESIUM ION, S-adenosylmethionine synthase, TRIPHOSPHATE, ... | | Authors: | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2021-07-21 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
6WPZ
 
 | | The structure of Pf4r from a superinfective isolate of the filamentous phage Pf4 of Pseudomonas aeruginosa PA01 | | Descriptor: | CHLORIDE ION, Pf4r | | Authors: | Michie, K.A, Norrian, P, Duggin, I.G, McDougald, D, Rice, S.A. | | Deposit date: | 2020-04-28 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | The Repressor C Protein, Pf4r, Controls Superinfection of Pseudomonas aeruginosa PAO1 by the Pf4 Filamentous Phage and Regulates Host Gene Expression.
Viruses, 13, 2021
|
|
7P8M
 
 | | Crystal structure of L147A/I351A variant of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii in complex with DMNB-SAM (4,5-dimethoxy-2-nitro benzyme S-adenosyl-methionine) | | Descriptor: | 4,5-dimethoxy-2-nitro benzyme S-adenosyl-methionine, MAGNESIUM ION, S-adenosylmethionine synthase, ... | | Authors: | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2021-07-23 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
7P82
 
 | | Crystal structure of apo form L147A/I351A variant of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii | | Descriptor: | S-adenosylmethionine synthase | | Authors: | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2021-07-21 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
6WHO
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
8DM5
 
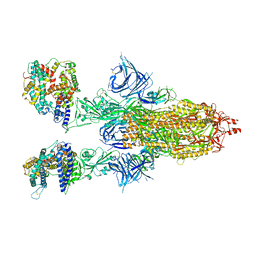 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
1CGE
 
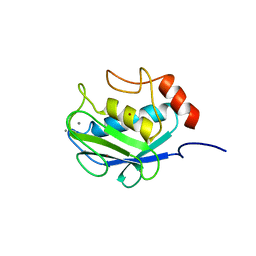 | | CRYSTAL STRUCTURES OF RECOMBINANT 19-KDA HUMAN FIBROBLAST COLLAGENASE COMPLEXED TO ITSELF | | Descriptor: | CALCIUM ION, FIBROBLAST COLLAGENASE, ZINC ION | | Authors: | Lovejoy, B, Hassell, A.M, Luther, M.A, Weigl, D, Jordan, S.R. | | Deposit date: | 1994-02-03 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of recombinant 19-kDa human fibroblast collagenase complexed to itself.
Biochemistry, 33, 1994
|
|
6WHN
 
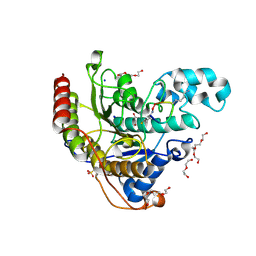 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHQ
 
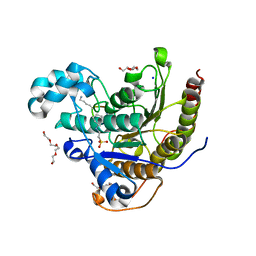 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
8B02
 
 | | Crystal structure of the dsRBD domain of tRNA-dihydrouridine(20) synthase from Amphimedon queenslandica | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, DRBM domain-containing protein, ... | | Authors: | Pecqueur, L, Faivre, B, Hamdane, D. | | Deposit date: | 2022-09-07 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.676 Å) | | Cite: | Evolutionary Diversity of Dus2 Enzymes Reveals Novel Structural and Functional Features among Members of the RNA Dihydrouridine Synthases Family.
Biomolecules, 12, 2022
|
|
