2FUV
 
 | | Phosphoglucomutase from Salmonella typhimurium. | | Descriptor: | MAGNESIUM ION, phosphoglucomutase | | Authors: | Osipiuk, J, Wu, R, Holzle, D, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-27 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of phosphoglucomutase from Salmonella typhimurium.
To be Published
|
|
2G59
 
 | |
4V59
 
 | | Crystal structure of fatty acid synthase complexed with nadp+ from thermomyces lanuginosus at 3.1 angstrom resolution. | | Descriptor: | FATTY ACID SYNTHASE ALPHA SUBUNITS, FATTY ACID SYNTHASE BETA SUBUNITS, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Jenni, S, Leibundgut, M, Boehringer, D, Frick, C, Mikolasek, B, Ban, N. | | Deposit date: | 2007-03-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Fungal Fatty Acid Synthase and Implications for Iterative Substrate Shuttling
Science, 316, 2007
|
|
2FN4
 
 | | The crystal structure of human Ras-related protein, RRAS, in the GDP-bound state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein R-Ras | | Authors: | Turnbull, A.P, Elkins, J.M, Gileadi, C, Burgess, N, Salah, E, Papagrigoriou, E, Debreczeni, J, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of human Ras-related protein, RRAS, in the GDP-bound state
To be Published
|
|
2FNB
 
 | | NMR STRUCTURE OF THE FIBRONECTIN ED-B DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (FIBRONECTIN) | | Authors: | Fattorusso, R, Pellecchia, M, Viti, F, Neri, P, Neri, D, Wuthrich, K. | | Deposit date: | 1998-12-16 | | Release date: | 1998-12-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the human oncofoetal fibronectin ED-B domain, a specific marker for angiogenesis.
Structure Fold.Des., 7, 1999
|
|
4UWF
 
 | | Discovery of (2S)-8-((3R)-3-Methylmorpholin-4-yl)-1-(3-methyl-2-oxo- butyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido(1,2-a)pyrimidin-6- one: a Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors | | Descriptor: | (8S)-9-[3,5-bis(fluoranyl)phenyl]-2-morpholin-4-yl-8-(trifluoromethyl)-7,8-dihydro-6H-pyrimido[1,2-a]pyrimidin-4-one, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT TYPE 3 | | Authors: | Pasquier, B, El-Ahmad, Y, Filoche-Romme, B, Dureuil, C, Fassy, F, Abecassis, P.Y, Mathieu, M, Bertrand, T, Benard, T, Barriere, C, ElBatti, S, Letallec, J.P, Sonnefraud, V, Brollo, M, Delbarre, L, Loyau, V, Pilorge, F, Bertin, L, Richepin, P, Arigon, J, Labrosse, J.R, Clement, J, Durand, F, Combet, R, Perraut, P, Leroy, V, Gay, F, Lefrancois, D, Bretin, F, Marquette, J.P, Michot, N, Caron, A, Castell, C, Schio, L, McCort, G, Goulaouic, H, Garcia-Echeverria, C, Ronan, B. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-Yl]-1-(3-Methyl-2-Oxobutyl)-2-(Trifluoromethyl)-3,4-Dihydro-2H-Pyrimido[1,2-A]Pyrimidin-6-One: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors.
J.Med.Chem., 58, 2015
|
|
2FUE
 
 | | Human alpha-Phosphomannomutase 1 with D-mannose 1-phosphate and Mg2+ cofactor bound | | Descriptor: | 1-O-phosphono-alpha-D-mannopyranose, MAGNESIUM ION, Phosphomannomutase 1 | | Authors: | Silvaggi, N.R, Zhang, C, Lu, Z, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2006-01-26 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-ray crystal structures of human alpha-phosphomannomutase 1 reveal the structural basis of congenital disorder of glycosylation type 1a.
J.Biol.Chem., 281, 2006
|
|
2FUQ
 
 | | Crystal Structure of Heparinase II | | Descriptor: | FORMIC ACID, PHOSPHATE ION, ZINC ION, ... | | Authors: | Shaya, D, Cygler, M. | | Deposit date: | 2006-01-27 | | Release date: | 2006-04-18 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Heparinase II from Pedobacter heparinus and Its Complex with a Disaccharide Product.
J.Biol.Chem., 281, 2006
|
|
4UYR
 
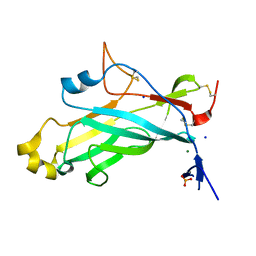 | | X-ray structure of the N-terminal domain of the flocculin Flo11 from Saccharomyces cerevisiae | | Descriptor: | FLOCCULATION PROTEIN FLO11, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Veelders, M, Kraushaar, T, Brueckner, S, Rhinow, D, Moesch, H.U, Essen, L.O. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-12 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-Like Adhesin Domain Girdled by Aromatic Bands.
Structure, 23, 2015
|
|
3UIN
 
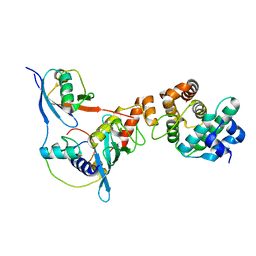 | | Complex between human RanGAP1-SUMO2, UBC9 and the IR1 domain from RanBP2 | | Descriptor: | E3 SUMO-protein ligase RanBP2, Ran GTPase-activating protein 1, SUMO-conjugating enzyme UBC9, ... | | Authors: | Gareau, J.R, Reverter, D, Lima, C.D. | | Deposit date: | 2011-11-05 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2.
J.Biol.Chem., 287, 2012
|
|
4V8T
 
 | | Cryo-EM Structure of the 60S Ribosomal Subunit in Complex with Arx1 and Rei1 | | Descriptor: | 25S RIBOSOMAL RNA, 5.8S RIBOSOMAL RNA, 5S RIBOSOMAL RNA, ... | | Authors: | Greber, B.J, Boehringer, D, Montellese, C, Ban, N. | | Deposit date: | 2012-08-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Cryo-Em Structures of Arx1 and Maturation Factors Rei1 and Jjj1 Bound to the 60S Ribosomal Subunit
Nat.Struct.Mol.Biol., 19, 2012
|
|
2FYU
 
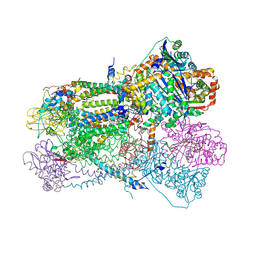 | | Crystal structure of bovine heart mitochondrial bc1 with jg144 inhibitor | | Descriptor: | (5S)-3-ANILINO-5-(2,4-DIFLUOROPHENYL)-5-METHYL-1,3-OXAZOLIDINE-2,4-DIONE, Cytochrome b, Cytochrome c1, ... | | Authors: | Xia, D, Esser, L. | | Deposit date: | 2006-02-08 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Surface-modulated motion switch: Capture and release of iron-sulfur protein in the cytochrome bc1 complex.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4W5A
 
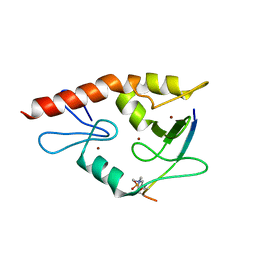 | | Complex structure of ATRX ADD bound to H3K9me3S10ph peptide | | Descriptor: | Peptide from Histone H3.3, Transcriptional regulator ATRX, ZINC ION | | Authors: | Zhao, D, Xiang, B, Li, H. | | Deposit date: | 2014-08-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ATRX tolerates activity-dependent histone H3 methyl/phos switching to maintain repetitive element silencing in neurons
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4V8L
 
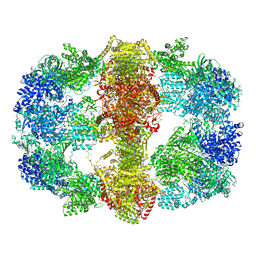 | |
2GAO
 
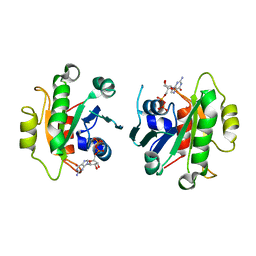 | | Crystal Structure of Human SAR1a in Complex With GDP | | Descriptor: | GTP-binding protein SAR1a, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Wang, J, Dimov, S, Tempel, W, Yaniw, D, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-09 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human SAR1a in Complex With GDP
To be Published
|
|
4WCE
 
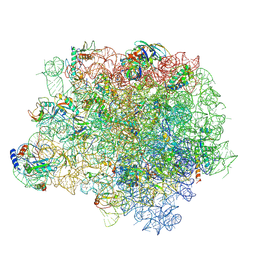 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S rRNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A. | | Deposit date: | 2014-09-04 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.526 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4V0U
 
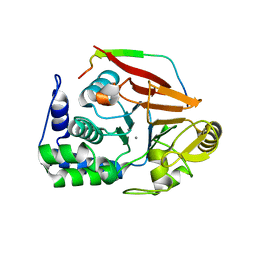 | | The crystal structure of ternary PP1G-PPP1R15B and G-actin complex | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, R, Yan, Y, Casado, A.C, Ron, D, Read, R.J. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (7.88 Å) | | Cite: | G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Elife, 4, 2015
|
|
4UWG
 
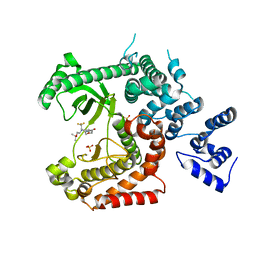 | | Discovery of (2S)-8-((3R)-3-Methylmorpholin-4-yl)-1-(3-methyl-2-oxo- butyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido(1,2-a)pyrimidin-6- one: a Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors | | Descriptor: | (8S)-2-(morpholin-4-yl)-9-[2-(propan-2-yloxy)ethyl]-8-(trifluoromethyl)-6,7,8,9-tetrahydro-4H-pyrimido[1,2-a]pyrimidin-4-one, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT TYPE 3, SULFATE ION | | Authors: | Pasquier, B, El-Ahmad, Y, Filoche-Romme, B, Dureuil, C, Fassy, F, Abecassis, P.Y, Mathieu, M, Bertrand, T, Benard, T, Barriere, C, ElBatti, S, Letallec, J.P, Sonnefraud, V, Brollo, M, Delbarre, L, Loyau, V, Pilorge, F, Bertin, L, Richepin, P, Arigon, J, Labrosse, J.R, Clement, J, Durand, F, Combet, R, Perraut, P, Leroy, V, Gay, F, Lefrancois, D, Bretin, F, Marquette, J.P, Michot, N, Caron, A, Castell, C, Schio, L, McCort, G, Goulaouic, H, Garcia-Echeverria, C, Ronan, B. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-Yl]-1-(3-Methyl-2-Oxobutyl)-2-(Trifluoromethyl)-3,4-Dihydro-2H-Pyrimido[1,2-A]Pyrimidin-6-One: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors.
J.Med.Chem., 58, 2015
|
|
4V3B
 
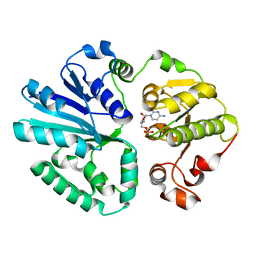 | | The structure of alpha2,3-sialyltransferase variant 1 from Pasteurella dagmatis in complex with the donor product CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
4UYS
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo11 from Saccharomyces cerevisiae | | Descriptor: | FLOCCULATION PROTEIN FLO11, MAGNESIUM ION, SODIUM ION | | Authors: | Kraushaar, T, Veelders, M, Brueckner, S, Rhinow, D, Moesch, H.U, Essen, L.O. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-12 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-Like Adhesin Domain Girdled by Aromatic Bands.
Structure, 23, 2015
|
|
4V4X
 
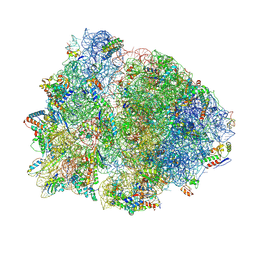 | | Crystal structure of the 70S Thermus thermophilus ribosome showing how the 16S 3'-end mimicks mRNA E and P codons. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Yusupova, G, Rees, B, Moras, D, Yusupov, M. | | Deposit date: | 2006-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Structural basis for messenger RNA movement on the ribosome.
Nature, 444, 2006
|
|
3UIP
 
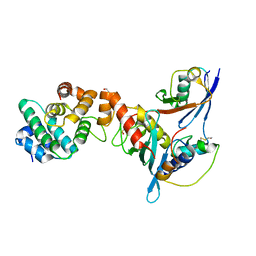 | | Complex between human RanGAP1-SUMO1, UBC9 and the IR1 domain from RanBP2 containing IR2 Motif II | | Descriptor: | E3 SUMO-protein ligase RanBP2, Ran GTPase-activating protein 1, SUMO-conjugating enzyme UBC9, ... | | Authors: | Gareau, J.R, Reverter, D, Lima, C.D. | | Deposit date: | 2011-11-05 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2.
J.Biol.Chem., 287, 2012
|
|
4V8N
 
 | | The crystal structure of agmatidine tRNA-Ile2 bound to the 70S ribosome in the A and P site. | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Mandal, D, Neubauer, C, Koehrer, C, RajBhandary, U.L, Ramakrishnan, V. | | Deposit date: | 2013-02-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Structural Basis for Specific Decoding of Aua by Isoleucine tRNA on the Ribosome
Nat.Struct.Mol.Biol., 20, 2013
|
|
2FVZ
 
 | | Human Inositol Monophosphosphatase 2 | | Descriptor: | Inositol monophosphatase 2 | | Authors: | Ogg, D, Hallberg, B.M, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Graslund, S, Hammarstrom, M, Hogbom, M, Holmberg-Schiavone, L, Kotenyova, T, Kursula, P, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Van Den Berg, S, Weigelt, J, Thorsell, A.G, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-21 | | Last modified: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Human Inositol Monophosphatase 2
To be published
|
|
4V1Y
 
 | | The structure of the hexameric atrazine chlorohydrolase, AtzA | | Descriptor: | 1,2-ETHANEDIOL, ATRAZINE CHLOROHYDROLASE, CHLORIDE ION, ... | | Authors: | Peat, T.S, Newman, J, Balotra, S, Lucent, D, Warden, A.C, Scott, C. | | Deposit date: | 2014-10-04 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of the Hexameric Atrazine Chlorohydrolase Atza.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
