6MPT
 
 | | TagT bound to LI-WTA | | Descriptor: | 2-(acetylamino)-2-deoxy-1-O-[(S)-{[(R)-{[(2Z,6Z,10E,14E,18E)-3,7,11,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaen-1-yl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-alpha-D-glucopyranose, CHLORIDE ION, Polyisoprenyl-teichoic acid--peptidoglycan teichoic acid transferase TagT | | Authors: | Owens, T.W, Schaefer, K, Kahne, D, Walker, S. | | Deposit date: | 2018-10-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Substrate Preferences Establish the Order of Cell Wall Assembly in Staphylococcus aureus.
J. Am. Chem. Soc., 140, 2018
|
|
1KMT
 
 | | Crystal structure of RhoGDI Glu(154,155)Ala mutant | | Descriptor: | Rho GDP-dissociation inhibitor 1 | | Authors: | Mateja, A, Devedjiev, Y, Krowarsh, D, Longenecker, K, Dauter, Z, Otlewski, J, Derewenda, Z.S. | | Deposit date: | 2001-12-17 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The impact of Glu-->Ala and Glu-->Asp mutations on the crystallization properties of RhoGDI: the structure of RhoGDI at 1.3 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2BGE
 
 | | Structure-based design of Protein Tyrosine Phosphatase-1B Inhibitors | | Descriptor: | 1,2,5-THIADIAZOLIDIN-3-ONE-1,1-DIOXIDE, PROTEIN-TYROSINE PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Black, E, Breed, J, Breeze, A.L, Embrey, K, Garcia, R, Gero, T.W, Godfrey, L, Kenny, P.W, Morley, A.D, Minshull, C.A, Pannifer, A.D, Read, J, Rees, A, Russell, D.J, Toader, D, Tucker, J. | | Deposit date: | 2004-12-21 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design of Protein Tyrosine Phosphatase-1B Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2BGD
 
 | | Structure-based design of Protein Tyrosine Phosphatase-1B Inhibitors | | Descriptor: | 5-(4-METHOXYBIPHENYL-3-YL)-1,2,5-THIADIAZOLIDIN-3-ONE 1,1-DIOXIDE, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Black, E, Breed, J, Breeze, A.L, Embrey, K, Garcia, R, Gero, T.W, Godfrey, L, Kenny, P.W, Morley, A.D, Minshull, C.A, Pannifer, A.D, Read, J, Rees, A, Russell, D.J, Toader, D, Tucker, J. | | Deposit date: | 2004-12-21 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of Protein Tyrosine Phosphatase-1B Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
6MTP
 
 | | Crystal structure of VRC42.04 Fab in complex with gp41 peptide | | Descriptor: | Antibody VRC42.04 Fab heavy chain, Antibody VRC42.04 Fab light chain, RV217 founder virus gp41 peptide | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Zhang, B, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.036 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
1KSU
 
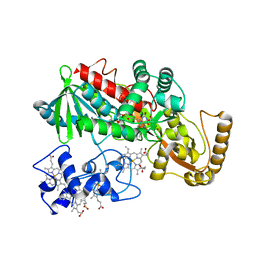 | | Crystal Structure of His505Tyr Mutant Flavocytochrome c3 from Shewanella frigidimarina | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FUMARIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pankhurst, K.L, Mowat, C.G, Miles, C.S, Leys, D, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2002-01-14 | | Release date: | 2002-08-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of His505 in the soluble fumarate reductase from Shewanella frigidimarina.
Biochemistry, 41, 2002
|
|
2BYO
 
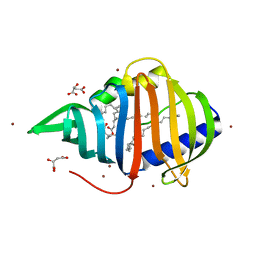 | | Crystal structure of Mycobacterium tuberculosis lipoprotein LppX (Rv2945c) | | Descriptor: | ACETATE ION, ALPHA-LINOLENIC ACID, D-MALATE, ... | | Authors: | Sulzenbacher, G, Canaan, S, Roig-Zamboni, V, Maurin, D, Gicquel, B, Bourne, Y. | | Deposit date: | 2005-08-03 | | Release date: | 2006-03-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Lppx is a Lipoprotein Required for the Translocation of Phthiocerol Dimycocerosates to the Surface of Mycobacterium Tuberculosis.
Embo J., 25, 2006
|
|
6MYY
 
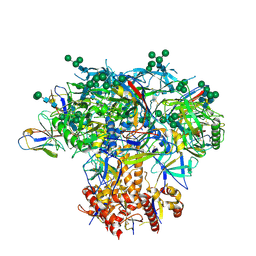 | | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer, 3 Fabs bound, sharpened map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 426c DS-SOSIP D3, ... | | Authors: | Borst, A.J, Weidle, C.E, Gray, M.D, Frenz, B, Snijder, J, Joyce, M.G, Georgiev, I.S, Stewart-Jones, G.B.E, Kwong, P.D, McGuire, A.T, DiMaio, F, Stamatatos, L, Pancera, M, Veesler, D. | | Deposit date: | 2018-11-02 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core.
Elife, 7, 2018
|
|
6N0O
 
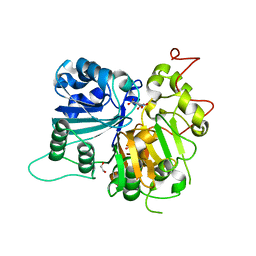 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ523 | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrobenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Crystal structure of Tdp1 catalytic domain
To Be Published
|
|
2BL5
 
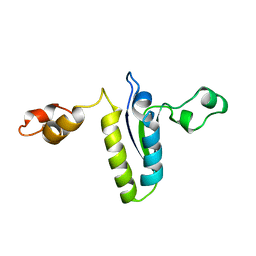 | | Solution structure of the KH-QUA2 region of the Xenopus STAR-GSG Quaking protein. | | Descriptor: | MGC83862 PROTEIN | | Authors: | Maguire, M.L, Guler-Gane, G, Nietlispach, D, Raine, A.R.C, Zorn, A.M, Standart, N, Broadhurst, R.W. | | Deposit date: | 2005-03-01 | | Release date: | 2005-04-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Kh-Qua2 Region of the Xenopus Star/Gsg Quaking Protein
J.Mol.Biol., 348, 2005
|
|
5OD3
 
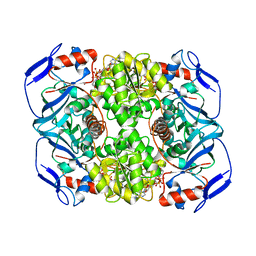 | | Crystal structure of R. ruber ADH-A, mutant Y54G, L119Y | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Stereo- and Regioselectivity in Catalyzed Transformation of a 1,2-Disubstituted Vicinal Diol and the Corresponding Diketone by Wild Type and Laboratory Evolved Alcohol Dehydrogenases
Acs Catalysis, 8, 2018
|
|
2BO5
 
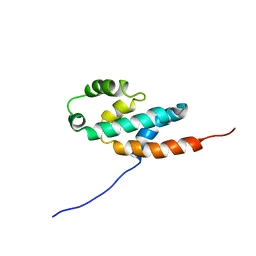 | | Bovine oligomycin sensitivity conferral protein N-terminal domain | | Descriptor: | ATP SYNTHASE OLIGOMYCIN SENSITIVITY CONFERRAL PROTEIN | | Authors: | Carbajo, R.J, Kellas, F.A, Runswick, M.J, Montgomery, M.G, Walker, J.E, Neuhaus, D. | | Deposit date: | 2005-04-07 | | Release date: | 2005-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the F1-binding domain of the stator of bovine F1Fo-ATPase and how it binds an alpha-subunit.
J. Mol. Biol., 351, 2005
|
|
6N4X
 
 | | Metabotropic Glutamate Receptor 5 Apo Form Ligand Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 5 | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6N2E
 
 | |
5MIH
 
 | | Crystal structure of the lectin LecA from Pseudomonas aeruginosa in complex with a phenyl-epoxy-galactopyranoside | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, PA-I galactophilic lectin, ... | | Authors: | Wagner, S, Hauk, D, Hofmann, M, Joachim, I, Sommer, R, Muller, R, Imberty, A, Varrot, A, Titz, A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent Lectin Inhibition and Application in Bacterial Biofilm Imaging.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2C9T
 
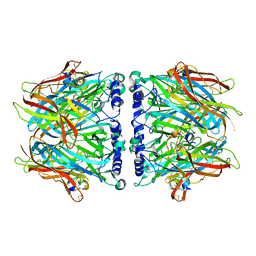 | | Crystal Structure Of Acetylcholine Binding Protein (AChBP) From Aplysia Californica In Complex With alpha-Conotoxin ImI | | Descriptor: | ALPHA-CONOTOXIN IMI, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Hogg, R.C, Celie, P.H, Bertrand, D, Tsetlin, V, Smit, A.B, Sixma, T.K. | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Determinants of Selective {Alpha}-Conotoxin Binding to a Nicotinic Acetylcholine Receptor Homolog Achbp.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
6MSP
 
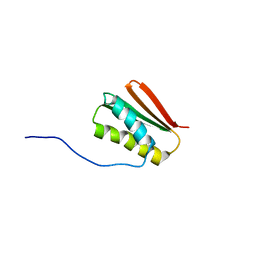 | | De novo Designed Protein Foldit3 | | Descriptor: | De novo Designed Protein Foldit3 | | Authors: | Liu, G, Ishida, Y, Swapna, G.V.T, Kleinfelter, S, Koepnick, B, Baker, D, Montelione, G.T. | | Deposit date: | 2018-10-17 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6N9L
 
 | | Crystal structure of T. maritima UvrA d117-399 with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, UvrABC system protein A, ZINC ION | | Authors: | Hartley, S, Case, B, Osuga, M, Hingorani, M.M, Jeruzalmi, D. | | Deposit date: | 2018-12-03 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The ATPase mechanism of UvrA2 reveals the distinct roles of proximal and distal ATPase sites in nucleotide excision repair.
Nucleic Acids Res., 47, 2019
|
|
2CGJ
 
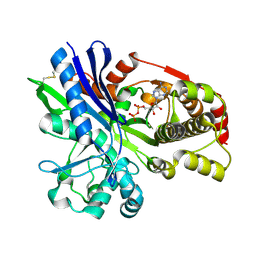 | |
2C7Q
 
 | |
1KUU
 
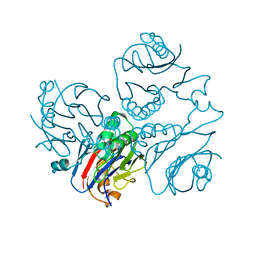 | | CRYSTAL STRUCTURE OF METHANOBACTERIUM THERMOAUTOTROPHICUM CONSERVED PROTEIN MTH1020 REVEALS AN NTN-HYDROLASE FOLD | | Descriptor: | conserved protein | | Authors: | Saridakis, V, Christendat, D, Thygesen, A, Arrowsmith, C.H, Edwards, A.M, Pai, E.F. | | Deposit date: | 2002-01-22 | | Release date: | 2002-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CRYSTAL STRUCTURE OF METHANOBACTERIUM THERMOAUTOTROPHICUM CONSERVED PROTEIN MTH1020 REVEALS AN NTN-HYDROLASE FOLD
PROTEINS: STRUCT.,FUNCT.,GENET., 48, 2002
|
|
2CIS
 
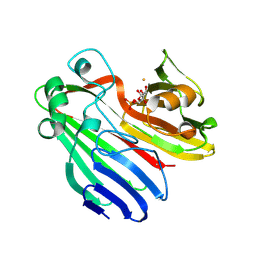 | | Structure-based functional annotation: Yeast ymr099c codes for a D- hexose-6-phosphate mutarotase. Complex with tagatose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-tagatofuranose, BARIUM ION, GLUCOSE-6-PHOSPHATE 1-EPIMERASE | | Authors: | Graille, M, Baltaze, J.-P, Leulliot, N, Liger, D, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2006-03-24 | | Release date: | 2006-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-based functional annotation: yeast ymr099c codes for a D-hexose-6-phosphate mutarotase.
J. Biol. Chem., 281, 2006
|
|
6MYZ
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ520 | | Descriptor: | 1,2-ETHANEDIOL, 4-oxo-8-phenyl-1,4-dihydroquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Structure of Tdp1 catalytic domain in complex with compound XZ520
To Be Published
|
|
2CD2
 
 | | LIGAND INDUCED CONFORMATIONAL CHANGES IN THE CRYSTAL STRUCTURES OF PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE COMPLEXES WITH FOLATE AND NADP+ | | Descriptor: | DIHYDROFOLATE REDUCTASE, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Rak, D, Luft, J, Pangborn, W, Queener, S. | | Deposit date: | 1999-03-15 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand-induced conformational changes in the crystal structures of Pneumocystis carinii dihydrofolate reductase complexes with folate and NADP+.
Biochemistry, 38, 1999
|
|
6MSQ
 
 | | Crystal structure of pRO-2.3 | | Descriptor: | pRO-2.3 | | Authors: | Boyken, S.E, Sankaran, B, Bick, M.J, Baker, D. | | Deposit date: | 2018-10-17 | | Release date: | 2019-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | De novo design of tunable, pH-driven conformational changes.
Science, 364, 2019
|
|
