9B1U
 
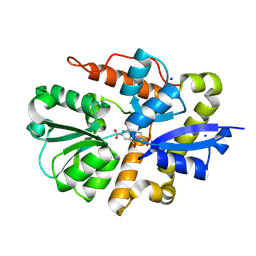 | | Crystal structure of PqqT with PQQ bound | | Descriptor: | PYRROLOQUINOLINE QUINONE, Putative ABC transporter periplasmic solute-binding protein, SODIUM ION | | Authors: | Boggs, D, Bruchs, A, Thompson, P, Olshansky, L, Bridwell-Rabb, J. | | Deposit date: | 2024-03-13 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-driven development of a biomimetic rare earth artificial metalloprotein.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
173L
 
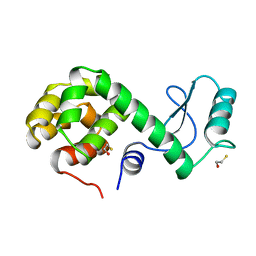 | | PROTEIN FLEXIBILITY AND ADAPTABILITY SEEN IN 25 CRYSTAL FORMS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Xiong, X.-P, Zhang, X.-J, Sun, D, Matthews, B.W. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
4V2W
 
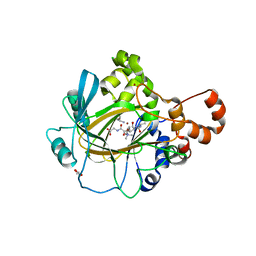 | | JMJD2A COMPLEXED WITH NI(II), NOG AND HISTONE H3K27me3 PEPTIDE (16-35) | | Descriptor: | CHLORIDE ION, GLYCEROL, HISTONE H3.1T, ... | | Authors: | Chowdhury, R, Zafred, D, Schofield, C.J. | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Studies on the Catalytic Domains of Multiple Jmjc Oxygenases Using Peptide Substrates.
Epigenetics, 9, 2014
|
|
171L
 
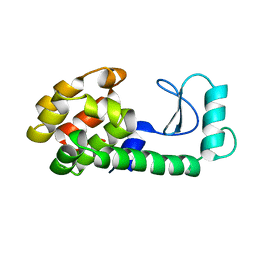 | |
1AB0
 
 | | C1G/V32D/F57H MUTANT OF MURINE ADIPOCYTE LIPID BINDING PROTEIN AT PH 4.5 | | Descriptor: | ADIPOCYTE LIPID BINDING PROTEIN | | Authors: | Ory, J, Kane, C, Simpson, M, Banaszak, L, Bernlohr, D. | | Deposit date: | 1997-01-30 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and crystallographic analyses of a portal mutant of the adipocyte lipid-binding protein.
J.Biol.Chem., 272, 1997
|
|
4X3X
 
 | | The crystal structure of Arc C-lobe | | Descriptor: | Activity-regulated cytoskeleton-associated protein | | Authors: | Zhang, W, Ward, M, Leahy, D, Worley, P. | | Deposit date: | 2014-12-01 | | Release date: | 2015-06-03 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of arc binding to synaptic proteins: implications for cognitive disease.
Neuron, 86, 2015
|
|
4WX2
 
 | | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with two F6F molecules in the alpha-site and one F6F molecule in the beta-site | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hilario, E, Caulkins, B.G, Young, R.P, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2014-11-13 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Visualizing the tunnel in tryptophan synthase with crystallography: Insights into a selective filter for accommodating indole and rejecting water.
Biochim.Biophys.Acta, 1864, 2016
|
|
4WW2
 
 | | Crystal structure of human TCR Alpha Chain-TRAV21-TRAJ8, Beta Chain-TRBV7-8, Antigen-presenting glycoprotein CD1d, and Beta-2-microglobulin | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Le Nours, J, Praveena, T, Pellicci, D, Gherardin, N.A, Lim, R.T, Besra, G, Keshipeddy, S, Richardson, S.K, Howell, A.R, Gras, S, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | Deposit date: | 2014-11-10 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
4X0V
 
 | | Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 | | Descriptor: | Beta-1,3-1,4-glucanase | | Authors: | Meng, D, Liu, X, Wang, X, Li, F, Feng, Y. | | Deposit date: | 2014-11-24 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Structural Insights into the Substrate Specificity of a Glycoside Hydrolase Family 5 Lichenase from Caldicellulosiruptor sp. F32
Biochem. J., 2017
|
|
4X3H
 
 | | CRYSTAL STRUCTURE OF ARC N-LOBE COMPLEXED WITH STARGAZIN PEPTIDE | | Descriptor: | Activity-regulated cytoskeleton-associated protein, VOLTAGE-DEPENDENT CALCIUM CHANNEL GAMMA-2 SUBUNIT | | Authors: | zhang, W, ward, m, leahy, d, worley, p. | | Deposit date: | 2014-11-30 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis of arc binding to synaptic proteins: implications for cognitive disease.
Neuron, 86, 2015
|
|
4X3I
 
 | | The crystal structure of Arc N-lobe complexed with CAMK2A fragment | | Descriptor: | Activity-regulated cytoskeleton-associated protein, CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE TYPE II SUBUNIT ALPHA | | Authors: | Zhang, W, Ward, M, Leahy, D, Worley, P. | | Deposit date: | 2014-11-30 | | Release date: | 2015-06-03 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of arc binding to synaptic proteins: implications for cognitive disease.
Neuron, 86, 2015
|
|
1C14
 
 | | CRYSTAL STRUCTURE OF E COLI ENOYL REDUCTASE-NAD+-TRICLOSAN COMPLEX | | Descriptor: | ENOYL REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Qiu, X, Janson, C, Court, R, Smyth, M, Payne, D, Abdel-Meguid, S. | | Deposit date: | 1999-07-20 | | Release date: | 2000-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for triclosan activity involves a flipping loop in the active site.
Protein Sci., 8, 1999
|
|
4XYN
 
 | | X-ray structure of Ca(2+)-S100B with human RAGE-derived W61 peptide | | Descriptor: | CALCIUM ION, Protein S100-B, Receptor for advanced glycation endproducts-derived peptide (W61) | | Authors: | Jensen, J.L, Indurthi, V.S.K, Neau, D, Vetter, S.W, Colbert, C.L. | | Deposit date: | 2015-02-02 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural insights into the binding of the human receptor for advanced glycation end products (RAGE) by S100B, as revealed by an S100B-RAGE-derived peptide complex.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1BL4
 
 | | FKBP MUTANT F36V COMPLEXED WITH REMODELED SYNTHETIC LIGAND | | Descriptor: | PROTEIN (FK506 BINDING PROTEIN), {3-[3-(3,4-DIMETHOXY-PHENYL)-1-(1-{1-[2-(3,4,5-TRIMETHOXY-PHENYL)-BUTYRYL]-PIPERIDIN-2YL}-VINYLOXY)-PROPYL]-PHENOXY}-ACETIC ACID | | Authors: | Hatada, M.H, Clackson, T, Yang, W, Rozamus, L.W, Amara, J, Rollins, C.T, Stevenson, L.F, Magari, S.R, Wood, S.A, Courage, N.L, Lu, X, Cerasoli Junior, F, Gilman, M, Holt, D. | | Deposit date: | 1998-07-23 | | Release date: | 1998-09-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4X9Z
 
 | | Dimeric conotoxin alphaD-GeXXA | | Descriptor: | alphaD-conotoxin GeXXA from the venom of Conus generalis | | Authors: | Xu, S, Zhang, T, Kompella, S, Adams, D, Ding, J, Wang, C. | | Deposit date: | 2014-12-12 | | Release date: | 2015-12-02 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conotoxin alpha D-GeXXA utilizes a novel strategy to antagonize nicotinic acetylcholine receptors
Sci Rep, 5, 2015
|
|
1B3J
 
 | | STRUCTURE OF THE MHC CLASS I HOMOLOG MIC-A, A GAMMADELTA T CELL LIGAND | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS I HOMOLOG MIC-A | | Authors: | Li, P, Willie, S, Bauer, S, Morris, D, Spies, T, Strong, R. | | Deposit date: | 1998-12-11 | | Release date: | 1999-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the MHC class I homolog MIC-A, a gammadelta T cell ligand.
Immunity, 10, 1999
|
|
1B0A
 
 | | 5,10, METHYLENE-TETRAHYDROPHOLATE DEHYDROGENASE/CYCLOHYDROLASE FROM E COLI. | | Descriptor: | PROTEIN (FOLD BIFUNCTIONAL PROTEIN) | | Authors: | Shen, B.W, Dyer, D, Huang, J.-Y, D'Ari, L, Rabinowitz, J, Stoddard, B.L. | | Deposit date: | 1998-11-06 | | Release date: | 1999-06-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The crystal structure of a bacterial, bifunctional 5,10 methylene-tetrahydrofolate dehydrogenase/cyclohydrolase.
Protein Sci., 8, 1999
|
|
1AKI
 
 | | THE STRUCTURE OF THE ORTHORHOMBIC FORM OF HEN EGG-WHITE LYSOZYME AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | LYSOZYME | | Authors: | Carter, D, He, J, Ruble, J.R, Wright, B. | | Deposit date: | 1997-05-19 | | Release date: | 1997-11-19 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structures of the Monoclinic and Orthorhombic Forms of Hen Egg-White Lysozyme at 6 Angstroms Resolution
Acta Crystallogr.,Sect.B, 38, 1982
|
|
4XJS
 
 | | Human CD38 complexed with inhibitor 1 [6-fluoro-2-methyl-4-[(2,3,6-trichlorobenzyl)amino]quinoline-8-carboxamide] | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, 6-fluoro-2-methyl-4-[(2,3,6-trichlorobenzyl)amino]quinoline-8-carboxamide, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1 | | Authors: | Shewchuk, L.M, Deaton, D, Stewart, E. | | Deposit date: | 2015-01-09 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 4-Amino-8-quinoline Carboxamides as Novel, Submicromolar Inhibitors of NAD-Hydrolyzing Enzyme CD38.
J.Med.Chem., 58, 2015
|
|
4Y32
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 109B with 14-3-3sigma | | Descriptor: | (2S)-2-(2-methoxyethyl)pyrrolidine, 14-3-3 protein sigma, ARG-THR-PRO-SEP-LEU-PRO-CNC(C(C)O)C(=O)N1CCCC1CCOC | | Authors: | Bartel, M, Milroy, L, Bier, D, Brunsveld, L, Ottmann, C. | | Deposit date: | 2015-02-10 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Stabilizer-Guided Inhibition of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1A8T
 
 | | METALLO-BETA-LACTAMASE IN COMPLEX WITH L-159,061 | | Descriptor: | 2-BUTYL-6-HYDROXY-3-[2'-(1H-TETRAZOL-5-YL)-BIPHENYL-4-YLMETHYL]-3H-QUINAZOLIN-4-ONE, METALLO-BETA-LACTAMASE, ZINC ION | | Authors: | Fitzgerald, P.M.D, Toney, J.H, Grover, N, Vanderwall, D. | | Deposit date: | 1998-03-23 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Antibiotic sensitization using biphenyl tetrazoles as potent inhibitors of Bacteroides fragilis metallo-beta-lactamase.
Chem.Biol., 5, 1998
|
|
1914
 
 | | SIGNAL RECOGNITION PARTICLE ALU RNA BINDING HETERODIMER, SRP9/14 | | Descriptor: | BETA-MERCAPTOETHANOL, PHOSPHATE ION, SIGNAL RECOGNITION PARTICLE 9/14 FUSION PROTEIN | | Authors: | Birse, D, Kapp, U, Strub, K, Cusack, S, Aberg, A. | | Deposit date: | 1997-11-13 | | Release date: | 1998-12-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The crystal structure of the signal recognition particle Alu RNA binding heterodimer, SRP9/14.
EMBO J., 16, 1997
|
|
4Y7I
 
 | | Crystal Structure of MTMR8 | | Descriptor: | Myotubularin-related protein 8, PHOSPHATE ION | | Authors: | Yoo, K, Lee, J, Son, J, Shin, W, Im, D, Heo, Y.S. | | Deposit date: | 2015-02-15 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of the catalytic phosphatase domain of MTMR8: implications for dimerization, membrane association and reversible oxidation.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1CDU
 
 | | STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4 MUTANT WITH PHE 43 REPLACED BY VAL | | Descriptor: | T-CELL SURFACE GLYCOPROTEIN CD4 | | Authors: | Wu, H, Myszka, D, Tendian, S.W, Brouillette, C.G, Sweet, R.W, Chaiken, I.M, Hendrickson, W.A. | | Deposit date: | 1996-11-11 | | Release date: | 1997-04-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Kinetic and structural analysis of mutant CD4 receptors that are defective in HIV gp120 binding.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
4YL2
 
 | | Aerococcus viridans L-lactate oxidase Y191F mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, Lactate oxidase, PYRUVIC ACID | | Authors: | Rainer, D, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2015-03-04 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Speeding up the product release: a second-sphere contribution from Tyr191 to the reactivity of l-lactate oxidase revealed in crystallographic and kinetic studies of site-directed variants.
Febs J., 282, 2015
|
|
