1Q35
 
 | | Crystal Structure of Pasteurella haemolytica Apo Ferric ion-Binding Protein A | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, iron binding protein FbpA | | Authors: | Shouldice, S.R, Dougan, D.R, Skene, R.J, Snell, G, Scheibe, D, Williams, P.A, Kirby, S, McRee, D.E, Schryvers, A.B, Tari, L.W. | | Deposit date: | 2003-07-28 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of Pasteurella haemolytica ferric ion-binding protein A reveals a novel class of bacterial iron-binding proteins
J.Biol.Chem., 278, 2003
|
|
2WUI
 
 | | Crystal Structure of MexZ, a key repressor responsible for antibiotic resistance in Pseudomonas aeruginosa. | | Descriptor: | TRANSCRIPTIONAL REGULATOR | | Authors: | Alguel, Y, Lu, D, Quade, N, Zhang, X. | | Deposit date: | 2009-10-05 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Mexz, a Key Repressor Responsible for Antibiotic Resistance in Pseudomonas Aeruginosa.
J.Struct.Biol., 172, 2010
|
|
8AZK
 
 | | Bovine 20S proteasome, untreated | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Szenkier, N, Arie, M, Matzov, D, Sertchook, R, Carmeli, R, Cascio, P, Stanhill, A, Shalev Benami, M, Navon, A. | | Deposit date: | 2022-09-06 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Bovine 20S proteasome, untreated
To be published
|
|
1PL1
 
 | | Crystal structure of human glutathione transferase (GST) A1-1 in complex with a decarboxy-glutathione | | Descriptor: | CHLORIDE ION, Glutathione S-transferase A1, N-(4-AMINOBUTANOYL)-S-(4-METHOXYBENZYL)-L-CYSTEINYLGLYCINE | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2X8F
 
 | | Native structure of Endo-1,5-alpha-L-arabinanases from Bacillus subtilis | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | de Sanctis, D, Inacio, J.M, Lindley, P.F, de Sa-Nogueira, I, Bento, I. | | Deposit date: | 2010-03-09 | | Release date: | 2011-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Evidence for the Role of Calcium in the Glycosidase Reaction of Gh43 Arabinanases.
FEBS J., 277, 2010
|
|
1Q1P
 
 | | E-Cadherin activation | | Descriptor: | CALCIUM ION, Epithelial-cadherin | | Authors: | Haussinger, D, Stetefeld, J. | | Deposit date: | 2003-07-22 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Proteolytic E-cadherin activation followed by solution NMR and X-ray crystallography.
Embo J., 23, 2004
|
|
2WM9
 
 | | Structure of the complex between DOCK9 and Cdc42. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GLYCEROL | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-06-30 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
2X98
 
 | | H.SALINARUM ALKALINE PHOSPHATASE | | Descriptor: | ALKALINE PHOSPHATASE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Wende, A, Johansson, P, Grininger, M, Oesterhelt, D. | | Deposit date: | 2010-03-14 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Characterization of a Halophilic Archaeal Alkaline Phosphatase.
J.Mol.Biol., 400, 2010
|
|
1Q2J
 
 | | Structural basis for tetrodotoxin-resistant sodium channel binding by mu-conotoxin SmIIIA | | Descriptor: | Mu-conotoxin SmIIIA | | Authors: | Keizer, D.W, West, P.J, Lee, E.F, Olivera, B.M, Bulaj, G, Yoshikami, D, Norton, R.S. | | Deposit date: | 2003-07-24 | | Release date: | 2004-02-24 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Structural basis for tetrodotoxin-resistant sodium channel binding by mu-conotoxin SmIIIA.
J.Biol.Chem., 278, 2003
|
|
8AND
 
 | | Domain swapped dimer of smolstatin (stefin) from Sphaerospora molnari | | Descriptor: | 1,2-ETHANEDIOL, Smolstatin | | Authors: | Havlickova, P, Bartosova-Sojkova, P, Sojka, D, Kascakova, B, Gavira, J.A, Kuta Smatanova, I. | | Deposit date: | 2022-08-05 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Domain swapped dimer of smolstatin (stefin) from Sphaerospora molnari
To Be Published
|
|
2WX0
 
 | | TAB2 NZF DOMAIN IN COMPLEX WITH Lys63-linked di-ubiquitin, P21 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 7-INTERACTING PROTEIN 2, UBIQUITIN, ZINC ION | | Authors: | Kulathu, Y, Akutsu, M, Bremm, A, Hofmann, K, Komander, D. | | Deposit date: | 2009-10-30 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two-Sided Ubiquitin Binding Explains Specificity of the Tab2 Nzf Domain
Nat.Struct.Mol.Biol., 16, 2009
|
|
8BTO
 
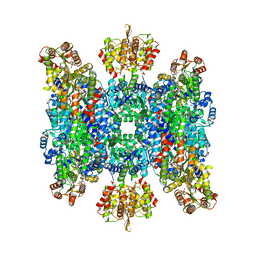 | | Helical structure of BcThsA in complex with 1''-3'gcADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, NAD(+) hydrolase ThsA, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tamulaitiene, G, Sasnauskas, G, Sabonis, D. | | Deposit date: | 2022-11-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Activation of Thoeris antiviral system via SIR2 effector filament assembly.
Nature, 627, 2024
|
|
1PQD
 
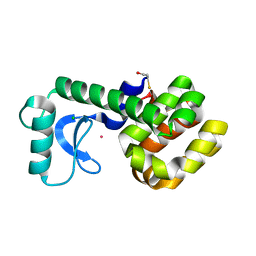 | | T4 LYSOZYME CORE REPACKING MUTANT CORE10/TA | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-06-18 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Repacking the Core of T4 Lysozyme by Automated Design
J.Mol.Biol., 332, 2003
|
|
2JB2
 
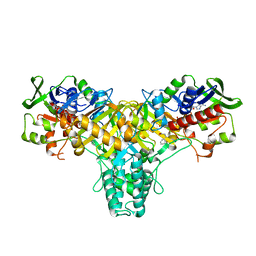 | | The structure of L-amino acid oxidase from Rhodococcus opacus in complex with L-phenylalanine. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE, PHENYLALANINE | | Authors: | Faust, A, Niefind, K, Hummel, W, Schomburg, D. | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation
J.Mol.Biol., 367, 2007
|
|
1PSY
 
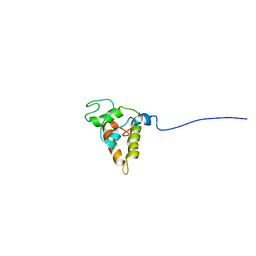 | | STRUCTURE OF RicC3, NMR, 20 STRUCTURES | | Descriptor: | 2S albumin | | Authors: | Pantoja-Uceda, D, Bruix, M, Gimenez-Gallego, G, Rico, M, Santoro, J. | | Deposit date: | 2003-06-22 | | Release date: | 2004-01-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RicC3, a 2S albumin storage protein from Ricinus communis.
Biochemistry, 42, 2003
|
|
2WIP
 
 | | STRUCTURE OF CDK2-CYCLIN A COMPLEXED WITH 8-ANILINO-1-METHYL-4,5-DIHYDRO- 1H-PYRAZOLO[4,3-H] QUINAZOLINE-3-CARBOXYLIC ACID | | Descriptor: | 1-methyl-8-(phenylamino)-4,5-dihydro-1H-pyrazolo[4,3-h]quinazoline-3-carboxylic acid, CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, ... | | Authors: | Brasca, M.G, Amboldi, N, Ballinari, D, Cameron, A.D, Casale, E, Cervi, G, Colombo, M, Colotta, F, Croci, V, Dalessio, R, Fiorentini, F, Isacchi, A, Mercurio, C, Moretti, W, Panzeri, A, Pastori, W, Pevarello, P, Quartieri, F, Roletto, F, Traquandi, G, Vianello, P, Vulpetti, A, Ciomei, M. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of N,1,4,4-Tetramethyl-8-{[4-(4-Methylpiperazin-1-Yl)Phenyl]Amino}-4,5-Dihydro-1H-Pyrazolo[4,3-H]Quinazoline-3-Carboxamide (Pha-848125), a Potent, Orally Available Cyclin Dependent Kinase Inhibitor.
J.Med.Chem., 52, 2009
|
|
2XKV
 
 | | Atomic Model of the SRP-FtsY Early Conformation | | Descriptor: | 4.5S RNA, CELL DIVISION PROTEIN FTSY, SIGNAL RECOGNITION PARTICLE PROTEIN | | Authors: | Estrozi, L.F, Boehringer, D, Shan, S.-o, Ban, N, Schaffitzel, C. | | Deposit date: | 2010-07-13 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Cryo-Em Structure of the E. Coli Translating Ribosome in Complex with Srp and its Receptor.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1PUX
 
 | | NMR Solution Structure of BeF3-Activated Spo0F, 20 conformers | | Descriptor: | Sporulation initiation phosphotransferase F | | Authors: | Gardino, A.K, Volkman, B.F, Cho, H.S, Lee, S.Y, Wemmer, D.E, Kern, D. | | Deposit date: | 2003-06-25 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of BeF(3)(-)-activated Spo0F reveals the conformational switch in a phosphorelay system.
J.Mol.Biol., 331, 2003
|
|
1NXM
 
 | | The high resolution structures of RmlC from Streptococcus suis | | Descriptor: | dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase | | Authors: | Dong, C, Major, L.L, Allen, A, Blankenfeldt, W, Maskell, D, Naismith, J.H. | | Deposit date: | 2003-02-11 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-Resolution Structures of RmlC from Streptococcus suis in Complex with Substrate Analogs Locate the Active Site of This Class of Enzyme
Structure, 11, 2003
|
|
2XB6
 
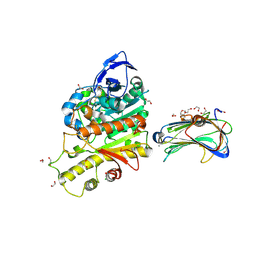 | | Revisited crystal structure of Neurexin1beta-Neuroligin4 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Leone, P, Comoletti, D, Ferracci, G, Conrod, S, Garcia, S.U, Taylor, P, Bourne, Y, Marchot, P. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights Into the Exquisite Selectivity of Neurexin-Neuroligin Synaptic Interactions
Embo J., 29, 2010
|
|
2XK5
 
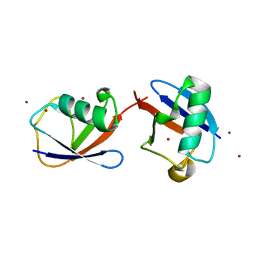 | |
2XCN
 
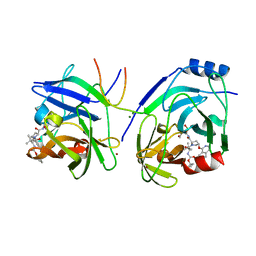 | | Crystal structure of HCV NS3 protease with a boronate inhibitor | | Descriptor: | MAGNESIUM ION, N-[(CYCLOPENTYLOXY)CARBONYL]-3-METHYL-L-VALYL-(4R)-N-{(1R)-3-HYDROXY-1-[HYDROXY(OXIDO)BORANYL]PROPYL}-4-(ISOQUINOLIN-1-YLOXY)-L-PROLINAMIDE, NS3 PROTEASE, ... | | Authors: | Li, X, Zhang, Y.-K, Liu, Y, Ding, C.Z, Li, Q, Zhou, Y, Plattner, J.J, Baker, S.J, Qian, X, Fan, D, Liao, L, Ni, Z.-J, White, G.V, Mordaunt, J.E, Lazarides, L.X, Slater, M.J, Jarvest, R.L, Thommes, P, Ellis, M, Edge, C.M, Hubbard, J.A, Nassau, P, McDowell, B, Skarzynski, T.J, Rowland, P, Somers, D.O, Kazmierski, W.M, Grimes, R.M, Wright, L.L, Smith, G.K, Zou, W, Wright, J, Pennicott, L.E. | | Deposit date: | 2010-04-23 | | Release date: | 2010-06-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Synthesis and Evaluation of Novel Alpha-Amino Cyclic Boronates as Inhibitors of Hcv Ns3 Protease.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XDQ
 
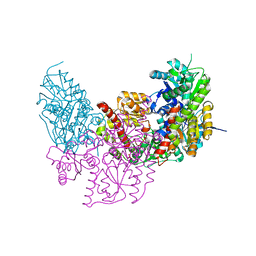 | | Dark Operative Protochlorophyllide Oxidoreductase (ChlN-ChlB)2 Complex | | Descriptor: | 1-METHYLGUANIDINE, IRON/SULFUR CLUSTER, LIGHT-INDEPENDENT PROTOCHLOROPHYLLIDE REDUCTASE SUBUNIT B, ... | | Authors: | Broecker, M.J, Schomburg, S, Heinz, D.W, Jahn, D, Schubert, W.-D, Moser, J. | | Deposit date: | 2010-05-06 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Nitrogenase-Like Dark Operative Protochlorophyllide Oxidoreductase Catalytic Complex (Chln/Chlb)2.
J.Biol.Chem., 285, 2010
|
|
1NYR
 
 | | Structure of Staphylococcus aureus threonyl-tRNA synthetase complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, THREONINE, ZINC ION, ... | | Authors: | Torres-Larios, A, Sankaranarayanan, R, Rees, B, Dock-Bregeon, A.C, Moras, D. | | Deposit date: | 2003-02-13 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational movements and cooperativity upon amino acid, ATP and tRNA binding in threonyl-tRNA synthetase
J.Mol.Biol., 331, 2003
|
|
8BH5
 
 | | SARS-CoV-2 BA.2.12.1 RBD in complex with Beta-27 Fab and C1 nanobody | | Descriptor: | Beta-27 heavy chain, Beta-27 light chain, GLYCEROL, ... | | Authors: | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Humoral responses against SARS-CoV-2 Omicron BA.2.11, BA.2.12.1 and BA.2.13 from vaccine and BA.1 serum.
Cell Discov, 8, 2022
|
|
