7A4M
 
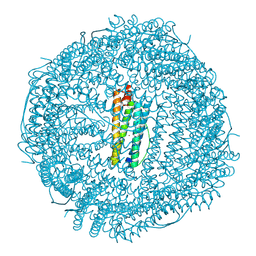 | | Cryo-EM structure of mouse heavy-chain apoferritin at 1.22 A | | Descriptor: | FE (III) ION, Ferritin heavy chain, ZINC ION | | Authors: | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (1.22 Å) | | Cite: | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
5VFQ
 
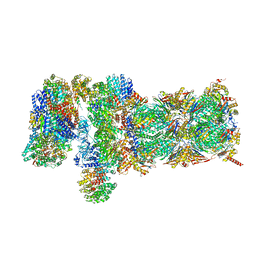 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
6ZML
 
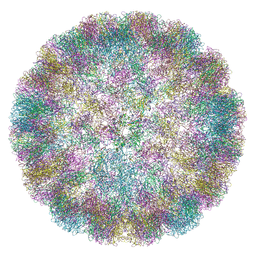 | | CryoEM Structure of Merkel Cell Polyomavirus Virus-like Particle | | Descriptor: | Capsid protein VP1 | | Authors: | Bayer, N.J, Januliene, D, Stehle, T, Moeller, A, Blaum, B.S. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-05 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of Merkel Cell Polyomavirus Capsid and Interaction with Its Glycosaminoglycan Attachment Receptor.
J.Virol., 94, 2020
|
|
5VHP
 
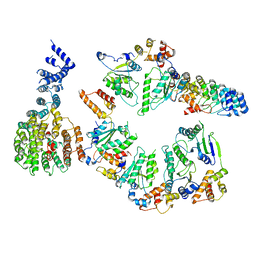 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
1W9N
 
 | | Isolation and characterization of epilancin 15X, a novel antibiotic from a clinical strain of Staphylococcus epidermidis | | Descriptor: | EPILANCIN 15X | | Authors: | Ekkelenkamp, M, Hanssen, M.G.M, Hsu, S.-T.D, de Jong, A, Milatovic, D, Verhoef, J, van Nuland, N.A.J. | | Deposit date: | 2004-10-14 | | Release date: | 2005-04-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Isolation and structural characterization of epilancin 15X, a novel lantibiotic from a clinical strain of Staphylococcus epidermidis.
FEBS Lett., 579, 2005
|
|
1W2G
 
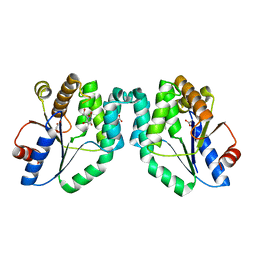 | | Crystal Structure Of Mycobacterium Tuberculosis Thymidylate Kinase Complexed With Deoxythymidine (dT) (2.1 A Resolution) | | Descriptor: | ACETATE ION, THYMIDINE, THYMIDYLATE KINASE TMK | | Authors: | Fioravanti, E, Adam, V, Munier-Lehmann, H, Bourgeois, D. | | Deposit date: | 2004-07-06 | | Release date: | 2005-01-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Mycobacterium Tuberculosis Thymidylate Kinase in Complex with 3'-Azidodeoxythymidine Monophosphate Suggests a Mechanism for Competitive Inhibition
Biochemistry, 44, 2005
|
|
1W7A
 
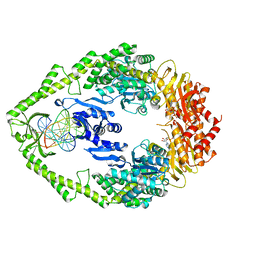 | | ATP bound MutS | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP* AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP* CP*T)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lamers, M.H, Georgijevic, D, Lebbink, J, Winterwerp, H.H.K, Agianian, B, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-08-31 | | Release date: | 2004-09-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | ATP Increases the Affinity between Muts ATPase Domains: Implications for ATP Hydrolysis and Conformational Changes
J.Biol.Chem., 279, 2004
|
|
5JP3
 
 | |
5VJQ
 
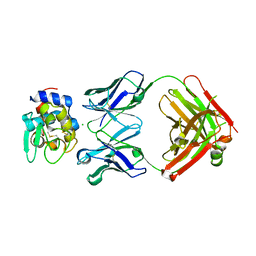 | | Complex between HyHEL10 Fab fragment heavy chain mutant (I29F, S52T, Y53F) and Pekin duck egg lysozyme isoform I (DEL-I) | | Descriptor: | CHLORIDE ION, GLYCEROL, HyHEL10 heavy chain Fab fragment carrying three mutations; I29F, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Germinal center antibody mutation trajectories are determined by rapid self/foreign discrimination.
Science, 360, 2018
|
|
1W90
 
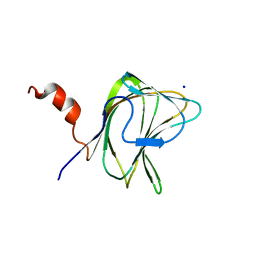 | | CBM29-2 mutant D114A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | 1,2-ETHANEDIOL, NON-CATALYTIC PROTEIN 1, SODIUM ION | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-10-01 | | Release date: | 2005-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1WBB
 
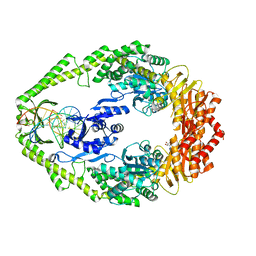 | | Crystal structure of E. coli DNA mismatch repair enzyme MutS, E38A mutant, in complex with a G.T mismatch | | Descriptor: | 5'-D(*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Georgijevic, D, Lebbink, J.H.G, Winterwerp, H.H.K, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-10-31 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dual Role of Muts Glutamate 38 in DNA Mismatch Discrimination and in the Authorization of Repair.
Embo J., 25, 2006
|
|
4IOS
 
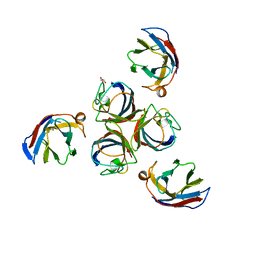 | | Structure of phage TP901-1 RBP (ORF49) in complex with nanobody 11. | | Descriptor: | BPP, GLYCEROL, Llama nanobody 11 | | Authors: | Desmyter, A, Farenc, C, Mahony, J, Spinelli, S, Bebeacua, C, Blangy, S, Veesler, D, van Sinderen, D, Cambillau, C. | | Deposit date: | 2013-01-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Viral infection modulation and neutralization by camelid nanobodies
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5W3Z
 
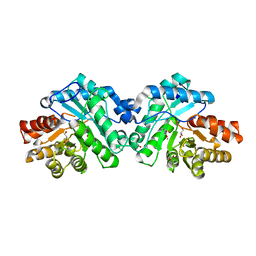 | | Crystal structure of SsoPox AsC6 mutant (L72I-Y99F-I122L-L228M-F229S-W263L) | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Hiblot, J, Gotthard, G, Jacquet, P, Daude, D, Bergonzi, C, Chabriere, E, Elias, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Rational engineering of a native hyperthermostable lactonase into a broad spectrum phosphotriesterase.
Sci Rep, 7, 2017
|
|
5W4U
 
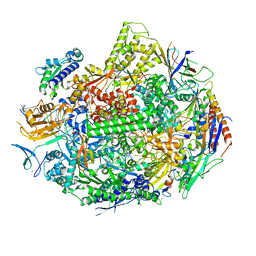 | |
6ZXX
 
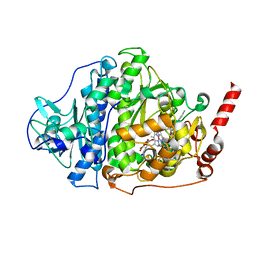 | | Catabolic reductive dehalogenase NpRdhA, N-terminally tagged. | | Descriptor: | 3 bromo 4 hydroxybenzoic acid, 3,5-bis(bromanyl)-4-oxidanyl-benzoic acid, BROMIDE ION, ... | | Authors: | Leys, D, Halliwell, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Catabolic Reductive Dehalogenase Substrate Complex Structures Underpin Rational Repurposing of Substrate Scope.
Microorganisms, 8, 2020
|
|
7ACA
 
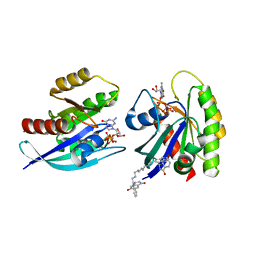 | | CRYSTAL STRUCTURE OF AN ACTIVE KRAS G12D (GPPCP) DIMER IN COMPLEX WITH BI-5747 | | Descriptor: | (3~{S})-5-oxidanyl-3-[2-[[6-[[3-[(1~{S})-6-oxidanyl-3-oxidanylidene-1,2-dihydroisoindol-1-yl]-1~{H}-indol-2-yl]methylamino]hexylamino]methyl]-1~{H}-indol-3-yl]-2,3-dihydroisoindol-1-one, 1,2-ETHANEDIOL, GTPase KRas, ... | | Authors: | Kessler, D. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ACTIVE KRAS G12D (GPPCP) DIMER IN COMPLEX WITH BI-5747
To Be Published
|
|
1W4W
 
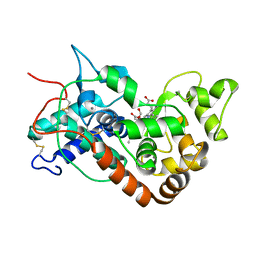 | | Ferric horseradish peroxidase C1A in complex with formate | | Descriptor: | CALCIUM ION, FORMIC ACID, HORSERADISH PEROXIDASE C1A, ... | | Authors: | Carlsson, G.H, Nicholls, P, Svistunenko, D, Berglund, G.I, Hajdu, J. | | Deposit date: | 2004-08-03 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Complexes of Horseradish Peroxidase with Formate, Acetate, and Carbon Monoxide
Biochemistry, 44, 2005
|
|
5JSL
 
 | | The L16F mutant of cytochrome c prime from Alcaligenes xylosoxidans: Ferrous form | | Descriptor: | ASCORBIC ACID, Cytochrome c', HEME C, ... | | Authors: | Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2016-05-08 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Engineering proximal vs. distal heme-NO coordination via dinitrosyl dynamics: implications for NO sensor design.
Chem Sci, 8, 2017
|
|
1W9F
 
 | | CBM29-2 mutant R112A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-10-12 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1WBD
 
 | | Crystal structure of E. coli DNA mismatch repair enzyme MutS, E38Q mutant, in complex with a G.T mismatch | | Descriptor: | 5'-D(*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Georgijevic, D, Lebbink, J.H.G, Winterwerp, H.H.K, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-10-31 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dual Role of Muts Glutamate 38 in DNA Mismatch Discrimination and in the Authorization of Repair.
Embo J., 25, 2006
|
|
6ZVO
 
 | | Crystal structure of unliganded MLKL executioner domain | | Descriptor: | BROMIDE ION, CHLORIDE ION, Mixed lineage kinase domain-like protein | | Authors: | Fiegen, D, Bauer, M, Nar, H. | | Deposit date: | 2020-07-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.371 Å) | | Cite: | Locking mixed-lineage kinase domain-like protein in its auto-inhibited state prevents necroptosis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1W54
 
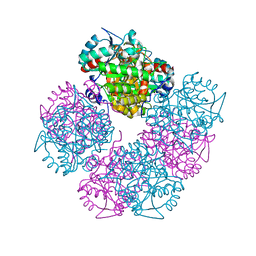 | | Stepwise introduction of a zinc binding site into Porphobilinogen synthase from Pseudomonas aeruginosa (mutation D139C) | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Frere, F, Reents, H, Schubert, W.-D, Heinz, D.W, Jahn, D. | | Deposit date: | 2004-08-05 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tracking the Evolution of Porphobilinogen Synthase Metal Dependence in Vitro
J.Mol.Biol., 345, 2005
|
|
7A4V
 
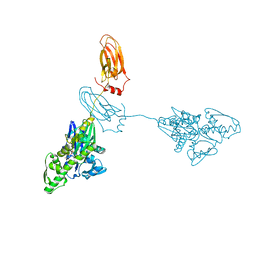 | |
5W72
 
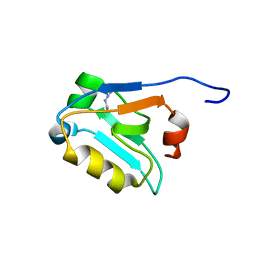 | |
8AJL
 
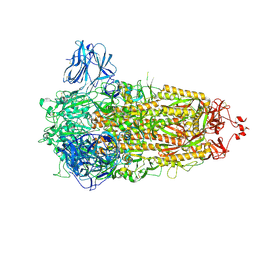 | | Structure of the Ancestral Scaffold Antigen-6 of Coronavirus Spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Hueting, D, Schriever, K, Wallden, K, Andrell, J, Syren, P.O. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Design, structure and plasma binding of ancestral beta-CoV scaffold antigens.
Nat Commun, 14, 2023
|
|
