2XIX
 
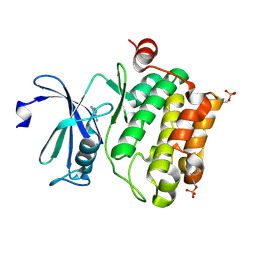 | | Protein kinase Pim-1 in complex with fragment-1 from crystallographic fragment screen | | Descriptor: | 3,5-DIAMINO-1H-[1,2,4]TRIAZOLE, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XJ2
 
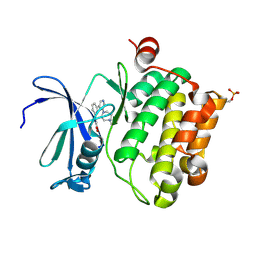 | | Protein kinase Pim-1 in complex with small molecule inhibitor | | Descriptor: | (2E)-3-{3-[6-(4-methyl-1,4-diazepan-1-yl)pyrazin-2-yl]phenyl}prop-2-enoic acid, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XIY
 
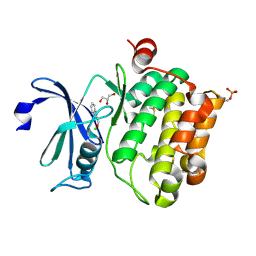 | | Protein kinase Pim-1 in complex with fragment-2 from crystallographic fragment screen | | Descriptor: | 2-HYDROXYMETHYL-BENZOIMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3HIZ
 
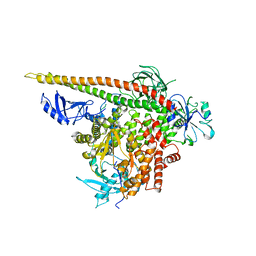 | | Crystal structure of p110alpha H1047R mutant in complex with niSH2 of p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Amzel, L.M, Vogelstein, B, Gabelli, S.B, Mandelker, D. | | Deposit date: | 2009-05-20 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A frequent kinase domain mutation that changes the interaction between PI3K{alpha} and the membrane.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2Y0D
 
 | | BceC mutation Y10K | | Descriptor: | SULFATE ION, UDP-GLUCOSE DEHYDROGENASE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Rocha, J, Popescu, A.O, Borges, P, Mil-Homens, D, Sa-Correia, I, Fialho, A.M, Frazao, C. | | Deposit date: | 2010-12-02 | | Release date: | 2011-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Burkholderia Cepacia Udp-Glucose Dehydrogenase (Ugd) Bcec and Role of Tyr10 in Final Hydrolysis of Ugd Thioester Intermediate.
J.Bacteriol., 193, 2011
|
|
2Y1X
 
 | | CRYSTAL STRUCTURE OF COACTIVATOR ASSOCIATED ARGININE METHYLTRANSFERASE 1 (CARM1) IN COMPLEX WITH SINEFUNGIN AND INDOLE INHIBITOR | | Descriptor: | CHLORIDE ION, HISTONE-ARGININE METHYLTRANSFERASE CARM1, N-(3-{5-[5-(1H-INDOL-4-YL)-1,3,4-OXADIAZOL-2-YL]-3-(TRIFLUOROMETHYL)-1H-PYRAZOL-1-YL}BENZYL)-L-ALANINAMIDE, ... | | Authors: | Sack, J.S, Thieffine, S, Bandiera, T, Fasolini, M, Duke, G.J, Jayaraman, L, Kish, K.F, Klei, H.E, Purandare, A.V, Rosettani, P, Troiani, S, Xie, D, Bertrand, J.A. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Carm1 Inhibition by Indole and Pyrazole Inhibitors
Biochem.J., 436, 2011
|
|
2XIT
 
 | | Crystal structure of monomeric MipZ | | Descriptor: | MIPZ | | Authors: | Kiekebusch, D, Michie, K.A, Essen, L.O, Lowe, J, Thanbichler, M. | | Deposit date: | 2010-06-30 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Localized Dimerization and Nucleoid Binding Drive Gradient Formation by the Bacterial Cell Division Inhibitor Mipz.
Mol.Cell, 46, 2012
|
|
3HHM
 
 | | Crystal structure of p110alpha H1047R mutant in complex with niSH2 of p85alpha and the drug wortmannin | | Descriptor: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Amzel, L.M, Vogelstein, B, Gabelli, S.B, Mandelker, D. | | Deposit date: | 2009-05-15 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A frequent kinase domain mutation that changes the interaction between PI3K{alpha} and the membrane.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2XJZ
 
 | | Crystal structure of the LMO2:LDB1-LID complex, C2 crystal form | | Descriptor: | CHLORIDE ION, LIM DOMAIN-BINDING PROTEIN 1, RHOMBOTIN-2, ... | | Authors: | El Omari, K, Karia, D, Porcher, C, Mancini, E.J. | | Deposit date: | 2010-07-06 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Leukemia Oncogene Lmo2: Implications for the Assembly of a Hematopoietic Transcription Factor Complex.
Blood, 117, 2011
|
|
2XTB
 
 | | Crystal Structure of Trypanosoma brucei rhodesiense Adenosine Kinase Complexed with Activator | | Descriptor: | 4-[5-(4-PHENOXYPHENYL)-1H-PYRAZOL-3-YL]MORPHOLINE, ADENOSINE KINASE | | Authors: | Kuettel, S, Greenwald, J, Kostrewa, D, Ahmed, S, Scapozza, L, Perozzo, R. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of T. B. Rhodesiense Adenosine Kinase Complexed with Inhibitor and Activator: Implications for Catalysis and Hyperactivation.
Plos Negl Trop Dis, 5, 2011
|
|
2Y5C
 
 | | Structure of human ferredoxin 2 (Fdx2)in complex with 2Fe2S cluster | | Descriptor: | ADRENODOXIN-LIKE PROTEIN, MITOCHONDRIAL, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Webert, H, Hobler, A, Sheftel, A.D, Molik, S, Maestre-Reyna, M, Essen, L.-O, Vorniscescu, D, Keusgen, M, Hannemann, F, Bernhardt, R, Lill, R. | | Deposit date: | 2011-01-12 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Functional Studies on Human Mitochondrial Ferredoxins
To be Published
|
|
3HF9
 
 | |
2Y4W
 
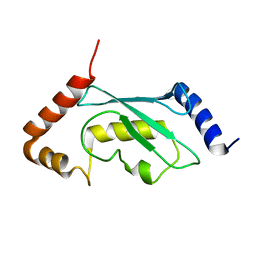 | | Solution structure of human ubiquitin conjugating enzyme Rad6b | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2 B | | Authors: | Huang, A, Hibbert, R.G, deJong, R.N, Das, D, Sixma, T.K, Boelens, R. | | Deposit date: | 2011-01-11 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Symmetry and Asymmetry of the Ring-Ring Dimer of Rad18
J.Mol.Biol., 410, 2011
|
|
3HF3
 
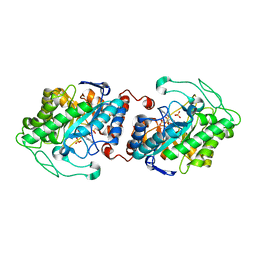 | | Old Yellow Enzyme from Thermus scotoductus SA-01 | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE, SULFATE ION | | Authors: | Opperman, D.J, Sewell, B.T, Litthauer, D, Isupov, M.N, Littlechild, J.A, van Heerden, E. | | Deposit date: | 2009-05-11 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a thermostable old yellow enzyme from Thermus scotoductus SA-01
Biochem.Biophys.Res.Commun., 393, 2010
|
|
2YQH
 
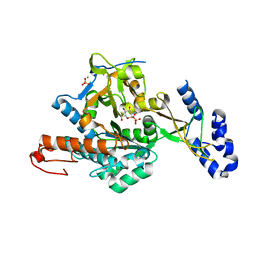 | | Crystal structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans, in the substrate-binding form | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Miki, K, Maruyama, D, Nishitani, Y, Nonaka, T, Kita, A. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Uridine-diphospho-N-acetylglucosamine Pyrophosphorylase from Candida albicans and Catalytic Reaction Mechanism
J.Biol.Chem., 282, 2007
|
|
2Y7Z
 
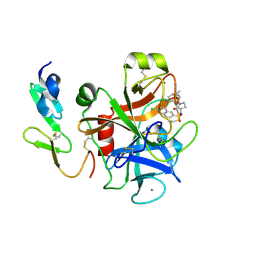 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-[(3S)-1-[(1S)-1-DIMETHYLAMINO-2,3-DIHYDRO-1H-INDEN-5-YL]-2-OXO-PYRROLIDIN-3-YL]NAPHTHALENE-2-SULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2YOM
 
 | | Solution NMR structure of the C-terminal extension of two bacterial light, oxygen, voltage (LOV) photoreceptor proteins from Pseudomonas putida | | Descriptor: | SENSORY BOX PROTEIN | | Authors: | Rani, R, Lecher, J, Hartmann, R, Krauss, U, Jaeger, K, Willbold, D. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-10 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Conservation of Dark Recovery Kinetic Parameters and Structural Features in the Pseudomonadaceae "Short" Light, Oxygen, Voltage (Lov) Protein Family: Implications for the Design of Lov-Based Optogenetic Tools.
Biochemistry, 52, 2013
|
|
2YOF
 
 | | Plasmodium falciparum thymidylate kinase in complex with a (thio)urea- beta-deoxythymidine inhibitor | | Descriptor: | 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-3-[[(2R,3S)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-oxidanyl-oxolan-2-yl]methyl]thiourea, ACETATE ION, THYMIDYLATE KINASE, ... | | Authors: | Huaqing, C, Carrero-Lerida, J, Silva, A.P.G, Whittingham, J.L, Brannigan, J.A, Ruiz-Perez, L.M, Read, K.D, Wilson, K.S, Gonzalez-Pacanowska, D, Gilbert, I.H. | | Deposit date: | 2012-10-24 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Synthesis and Evaluation of Alpha-Thymidine Analogues as Novel Antimalarials.
J.Med.Chem., 55, 2012
|
|
2YE0
 
 | | X-ray structure of the cyan fluorescent protein mTurquoise (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
3HJC
 
 | | Crystal structure of the carboxy-terminal domain of HSP90 from Leishmania major, LmjF33.0312 | | Descriptor: | Heat shock protein 83-1, SULFATE ION | | Authors: | Wernimont, A.K, Tempel, W, Walker, J, Lin, Y.H, Hutchinson, A, Mackenzie, F, Fairlamb, A, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the middle and carboxy-terminal domain of HSP90 from Leishmania major, LMJF33.0312
To be Published
|
|
2Y82
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-((3S)-2-OXO-1-{4-[(2S)-2-PYRROLIDINYL]PHENYL}-3-PYRROLIDINYL)-2-NAPHTHALENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2Y8R
 
 | | Crystal structure of apo AMA1 mutant (Tyr230Ala) from Toxoplasma gondii | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, APICAL MEMBRANE ANTIGEN, PUTATIVE | | Authors: | Tonkin, M.L, Roques, M, Lamarque, M.H, Pugniere, M, Douguet, D, Crawford, J, Lebrun, M, Boulanger, M.J. | | Deposit date: | 2011-02-10 | | Release date: | 2011-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Host Cell Invasion by Apicomplexan Parasites: Insights from the Co-Structure of Ama1 with a Ron2 Peptide
Science, 333, 2011
|
|
2YE1
 
 | | X-ray structure of the cyan fluorescent proteinmTurquoise-GL (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN, MAGNESIUM ION | | Authors: | von Stetten, D, Noirclerc-Savoye, M, Goedhart, J, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Characterization of the Cyan Fluorescent Protein Mturquoise-Gl
To be Published
|
|
3HKF
 
 | | Murine unglycosylated IgG Fc fragment | | Descriptor: | CHLORIDE ION, Igh protein, MAGNESIUM ION | | Authors: | Feige, M.J, Nath, S, Catharino, S.R, Weinfurtner, D, Steinbacher, S, Buchner, J. | | Deposit date: | 2009-05-23 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the murine unglycosylated IgG1 Fc fragment
J.Mol.Biol., 391, 2009
|
|
3HHA
 
 | | Crystal structure of cathepsin L in complex with AZ12878478 | | Descriptor: | ACETATE ION, Cathepsin L1, GLYCEROL, ... | | Authors: | Asaad, N, Bethel, P.A, Coulson, M.D, Dawson, J, Ford, S.J, Gerhardt, S, Grist, M, Hamlin, G.A, James, M.J, Jones, E.V, Karoutchi, G.I, Kenny, P.W, Morley, A.D, Oldham, K, Rankine, N, Ryan, D, Wells, S.L, Wood, L, Augustin, M, Krapp, S, Simader, H, Steinbacher, S. | | Deposit date: | 2009-05-15 | | Release date: | 2009-06-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Dipeptidyl nitrile inhibitors of Cathepsin L.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
