2VWY
 
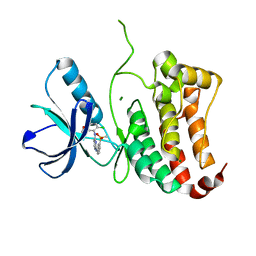 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, N'-(5-chloro-1,3-benzodioxol-4-yl)-N-(3-methylsulfonylphenyl)pyrimidine-2,4-diamine | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 2: Structure-Based Discovery and Optimisation of 3,5-Bis Substituted Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3AEM
 
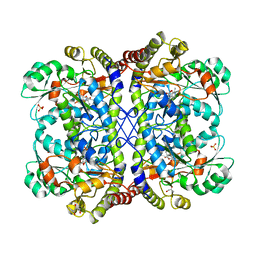 | | Reaction intermediate structure of Entamoeba histolytica methionine gamma-lyase 1 containing Michaelis complex and methionine imine-pyridoxamine-5'-phosphate | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-4-(methylsulfanyl)butanoic acid, GLYCEROL, METHIONINE, ... | | Authors: | Karaki, T, Sato, D, Shimizu, A, Nozaki, T, Harada, S. | | Deposit date: | 2010-02-10 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Entamoeba histolytica methionine gamma-lyase 1
To be published
|
|
2VTM
 
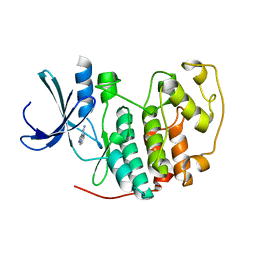 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, PYRAZOLO[1,5-A]PYRIMIDINE-3-CARBONITRILE | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VU3
 
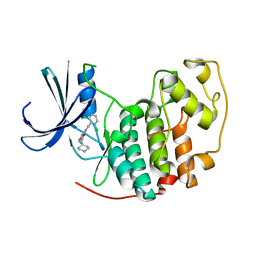 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | 4-{[(2,6-dichlorophenyl)carbonyl]amino}-N-piperidin-4-yl-1H-pyrazole-3-carboxamide, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-20 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
3A4N
 
 | | Crystal structure of archaeal O-phosphoseryl-tRNA(Sec) kinase | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, L-seryl-tRNA(Sec) kinase, ... | | Authors: | Araiso, Y, Ishitani, R, Soll, D, Nureki, O. | | Deposit date: | 2009-07-10 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a tRNA-dependent kinase essential for selenocysteine decoding
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3AEN
 
 | | Reaction intermediate structure of Entamoeba histolytica methionine gamma-lyase 1 containing Michaelis complex and alpha-amino-alpha, beta-butenoic acid-pyridoxal-5'-phosphate | | Descriptor: | (2E)-2-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}but-2-enoic acid, GLYCEROL, METHIONINE, ... | | Authors: | Karaki, T, Sato, D, Shimizu, A, Nozaki, T, Harada, S. | | Deposit date: | 2010-02-10 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Entamoeba histolytica methionine gamma-lyase 1
To be Published
|
|
3O73
 
 | | Crystal structure of quinone reductase 2 in complex with the indolequinone MAC627 | | Descriptor: | 5-[(4-aminobutyl)amino]-1,2-dimethyl-3-[(4-nitrophenoxy)methyl]-1H-indole-4,7-dione, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Dufour, M, Yan, C, Colucci, M.A, Siegel, D, Li, Y, De Matteis, C.I, Ross, D, Moody, C.J. | | Deposit date: | 2010-07-30 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism-Based Inhibition of Quinone Reductase 2 (NQO2): Selectivity for NQO2 over NQO1 and Structural Basis for Flavoprotein Inhibition.
Chembiochem, 12, 2011
|
|
4XFO
 
 | |
6PCW
 
 | | Human PIM1 bound to benzothiophene inhibitor 213 | | Descriptor: | 4-[5-(cyclopropylcarbamoyl)thiophen-2-yl]-1-benzothiophene-2-carboxamide, GLYCEROL, Peptide, ... | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-18 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PIM1 bound to benzothiophene inhibitor
To Be Published
|
|
8HT2
 
 | |
2YIN
 
 | | STRUCTURE OF THE COMPLEX BETWEEN Dock2 AND Rac1. | | Descriptor: | DEDICATOR OF CYTOKINESIS PROTEIN 2, RAS-RELATED C3 BOTULINUM TOXIN SUBSTRATE 1 | | Authors: | Kulkarni, K.A, Yang, J, Zhang, Z, Barford, D. | | Deposit date: | 2011-05-16 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Multiple Factors Confer Specific Cdc42 and Rac Protein Activation by Dedicator of Cytokinesis (Dock) Nucleotide Exchange Factors.
J.Biol.Chem., 286, 2011
|
|
8HIU
 
 | |
4XJE
 
 | |
4XFN
 
 | |
8HT4
 
 | |
2YMM
 
 | | Sulfate bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE, SULFATE ION | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-09 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
6PIL
 
 | | Antibody scFv-M204 monomeric state | | Descriptor: | CHLORIDE ION, scFv-M204 antibody | | Authors: | Abskharon, R, Sawaya, M.R, Seidler, P.M, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2019-06-26 | | Release date: | 2020-06-24 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a conformational antibody that binds tau oligomers and inhibits pathological seeding by extracts from donors with Alzheimer's disease.
J.Biol.Chem., 295, 2020
|
|
3DMK
 
 | | Crystal structure of Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, N-terminal eight Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, ... | | Authors: | Sawaya, M.R, Wojtowicz, W.M, Eisenberg, D, Zipursky, S.L. | | Deposit date: | 2008-07-01 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.19 Å) | | Cite: | A double S shape provides the structural basis for the extraordinary binding specificity of Dscam isoforms.
Cell(Cambridge,Mass.), 134, 2008
|
|
6P0Y
 
 | | Cryptosporidium parvum pyruvate kinase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2019-05-17 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An overview of structure, function, and regulation of pyruvate kinases.
Protein Sci., 28, 2019
|
|
2W0J
 
 | | Crystal structure of Chk2 in complex with NSC 109555, a specific inhibitor | | Descriptor: | 4,4'-DIACETYLDIPHENYLUREA-BIS(GUANYLHYDRAZONE), NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Tropea, J.E, Zhang, D, Jobson, A.G, Pommier, Y, Shoemaker, R.H, Waugh, D.S. | | Deposit date: | 2008-08-18 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Checkpoint Kinase 2 in Complex with Nsc 109555, a Potent and Selective Inhibitor
Protein Sci., 18, 2009
|
|
2YKT
 
 | | Crystal structure of the I-BAR domain of IRSp53 (BAIAP2) in complex with an EHEC derived Tir peptide | | Descriptor: | BRAIN-SPECIFIC ANGIOGENESIS INHIBITOR 1-ASSOCIATED PROTEIN 2, SULFATE ION, TRANSLOCATED INTIMIN RECEPTOR PROTEIN | | Authors: | de Groot, J.C, Schlueter, K, Carius, Y, Quedenau, C, Vingadassalom, D, Faix, J, Weiss, S.M, Reichelt, J, Standfuss-Gabisch, C, Lesser, C.F, Leong, J.M, Heinz, D.W, Buessow, K, Stradal, T.E.B. | | Deposit date: | 2011-05-30 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Basis for Complex Formation between Human Irsp53 and the Translocated Intimin Receptor Tir of Enterohemorrhagic E. Coli.
Structure, 19, 2011
|
|
4XNW
 
 | | The human P2Y1 receptor in complex with MRS2500 | | Descriptor: | P2Y purinoceptor 1,Rubredoxin,P2Y purinoceptor 1, ZINC ION, [(1R,2S,4S,5S)-4-[2-iodo-6-(methylamino)-9H-purin-9-yl]-2-(phosphonooxy)bicyclo[3.1.0]hex-1-yl]methyl dihydrogen phosphate | | Authors: | Zhang, D, Gao, Z, Jacobson, K, Han, G.W, Stevens, R, Zhao, Q, Wu, B, GPCR Network (GPCR) | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-01 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two disparate ligand-binding sites in the human P2Y1 receptor
Nature, 520, 2015
|
|
2YNF
 
 | | HIV-1 Reverse Transcriptase Y188L mutant in complex with inhibitor GSK560 | | Descriptor: | 2-azanyl-N-[[4-bromanyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-4-chloranyl-1H-imidazole-5-carboxamide, D(-)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-14 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
3DS6
 
 | | P38 complex with a phthalazine inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-cyclopropyl-4-methyl-3-[1-(2-methylphenyl)phthalazin-6-yl]benzamide | | Authors: | Herberich, B, Syed, R, Li, V, Grosfeld, D. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of highly selective and potent p38 inhibitors based on a phthalazine scaffold.
J.Med.Chem., 51, 2008
|
|
2YOH
 
 | | Plasmodium falciparum thymidylate kinase in complex with a urea-alpha- deoxythymidine inhibitor | | Descriptor: | 1-[[(2R,3S,5S)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-oxidanyl-oxolan-2-yl]methyl]-3-(4-nitrophenyl)urea, THYMIDYLATE KINASE | | Authors: | Huaqing, C, Carrero-Lerida, J, Silva, A.P.G, Whittingham, J.L, Brannigan, J.A, Ruiz-Perez, L.M, Read, K.D, Wilson, K.S, Gonzalez-Pacanowska, D, Gilbert, I.H. | | Deposit date: | 2012-10-24 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Synthesis and Evaluation of Alpha-Thymidine Analogues as Novel Antimalarials.
J.Med.Chem., 55, 2012
|
|
