7ZQ0
 
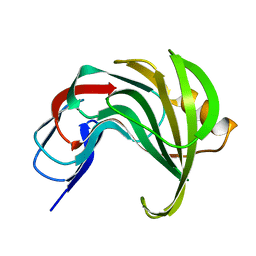 | | Room temperature SSX structure of GH11 xylanase from Nectria haematococca (1000 frames) | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Oberthuer, D, Andaleeb, H, Betzel, C, Perbandt, M, Yefanov, O, Zielinski, K. | | 登録日 | 2022-04-29 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
7ZRE
 
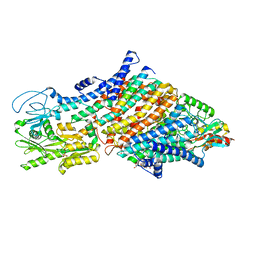 | | Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, under turnover conditions | | 分子名称: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
3ZUX
 
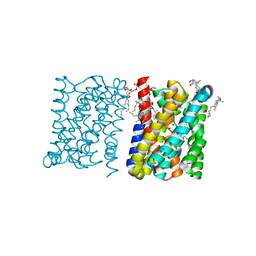 | | Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. | | 分子名称: | LAURYL DIMETHYLAMINE-N-OXIDE, MERCURY (II) ION, PHOSPHATIDYLETHANOLAMINE, ... | | 著者 | Hu, N.-J, Iwata, S, Cameron, A.D, Drew, D. | | 登録日 | 2011-07-21 | | 公開日 | 2011-10-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of a Bacterial Homologue of the Bile Acid Sodium Symporter Asbt.
Nature, 478, 2011
|
|
2JDT
 
 | | Structure of PKA-PKB chimera complexed with ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY) ETHYLAMINO)ETHYL)AMIDE | | 分子名称: | CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY)ETHYLAMINO)ETHYL)AMIDE | | 著者 | Davies, T.G, Verdonk, M.L, Graham, B, Saalau-Bethell, S, Hamlett, C.C.F, McHardy, T, Collins, I, Garrett, M.D, Workman, P, Woodhead, S.J, Jhoti, H, Barford, D. | | 登録日 | 2007-01-12 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | A Structural Comparison of Inhibitor Binding to Pkb, Pka and Pka-Pkb Chimera
J.Mol.Biol., 367, 2007
|
|
7ZRH
 
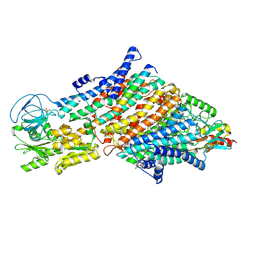 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | 分子名称: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRK
 
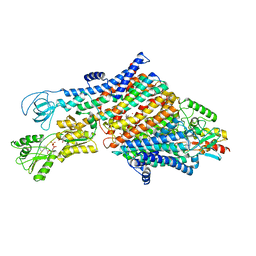 | | Cryo-EM map of the WT KdpFABC complex in the E1-P_ADP conformation, under turnover conditions | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
3ZUY
 
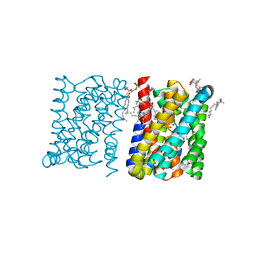 | | Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. | | 分子名称: | LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | 著者 | Hu, N.-J, Iwata, S, Cameron, A.D, Drew, D. | | 登録日 | 2011-07-21 | | 公開日 | 2011-10-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of a Bacterial Homologue of the Bile Acid Sodium Symporter Asbt.
Nature, 478, 2011
|
|
7ZQS
 
 | | Cryo-EM Structure of Human Transferrin Receptor 1 bound to DNA Aptamer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (30-MER), ... | | 著者 | Bansia, H, Wang, T, Gutierrez, D, des Georges, A. | | 登録日 | 2022-05-02 | | 公開日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (2.54 Å) | | 主引用文献 | Discovery of a Transferrin Receptor 1-Binding Aptamer and Its Application in Cancer Cell Depletion for Adoptive T-Cell Therapy Manufacturing.
J.Am.Chem.Soc., 144, 2022
|
|
3ZD0
 
 | | The Solution Structure of Monomeric Hepatitis C Virus p7 Yields Potent Inhibitors of Virion Release | | 分子名称: | P7 PROTEIN | | 著者 | Foster, T.L, Sthompson, G, Kalverda, A.P, Kankanala, J, Thompson, J, Barker, A.M, Clarke, D, Noerenberg, M, Pearson, A.R, Rowlands, D.J, Homans, S.W, Harris, M, Foster, R, Griffin, S.D.C. | | 登録日 | 2012-11-23 | | 公開日 | 2013-09-04 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-Guided Design Affirms Inhibitors of Hepatitis C Virus P7 as a Viable Class of Antivirals Targeting Virion Release
Hepatology, 59, 2014
|
|
7ZRJ
 
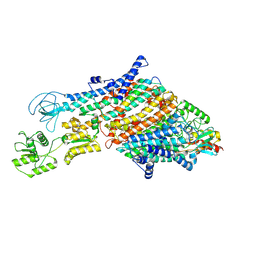 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | 分子名称: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZPV
 
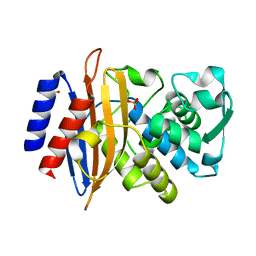 | | Room temperature SSX crystal structure of CTX-M-14 | | 分子名称: | Beta-lactamase, SULFATE ION | | 著者 | Oberthuer, D, Perbandt, M, Prester, A, Rohde, H, Betzel, C, Yefanov, O. | | 登録日 | 2022-04-29 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
2JIL
 
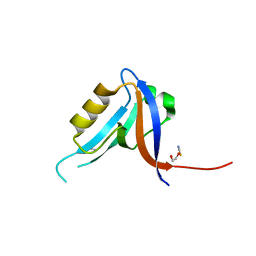 | | Crystal structure of 2nd PDZ domain of glutamate receptor interacting protein-1 (GRIP1) | | 分子名称: | 1,2-ETHANEDIOL, GLUTAMATE RECEPTOR INTERACTING PROTEIN-1, THIOCYANATE ION | | 著者 | Tickle, J, Elkins, J, Pike, A.C.W, Cooper, C, Salah, E, Papagrigoriou, E, von Delft, F, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D. | | 登録日 | 2007-06-28 | | 公開日 | 2007-07-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of 2Nd Pdz Domain of Glutamate Receptor Interacting Protein-1 (Grip1)
To be Published
|
|
7ZRG
 
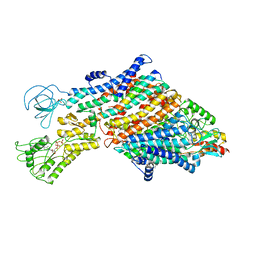 | | Cryo-EM map of the WT KdpFABC complex in the E1_ATPearly conformation, under turnover conditions | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Rheinberger, J, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
2JK2
 
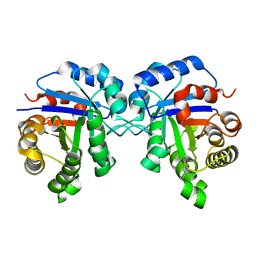 | | STRUCTURAL BASIS OF HUMAN TRIOSEPHOSPHATE ISOMERASE DEFICIENCY. CRYSTAL STRUCTURE OF THE WILD TYPE ENZYME. | | 分子名称: | TRIOSEPHOSPHATE ISOMERASE | | 著者 | Rodriguez-Almazan, C, Arreola-Alemon, R, Rodriguez-Larrea, D, Aguirre-Lopez, B, De Gomez-Puyou, M.T, Perez-Montfort, R, Costas, M, Gomez-Puyou, A, Torres-Larios, A. | | 登録日 | 2008-06-22 | | 公開日 | 2008-07-01 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Basis of Human Triosephosphate Isomerase Deficiency: Mutation E104D is Related to Alterations of a Conserved Water Network at the Dimer Interface.
J.Biol.Chem., 283, 2008
|
|
3DXN
 
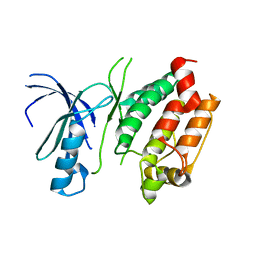 | | Crystal structure of the calcium-dependent kinase from toxoplasma gondii, 541.m00134, kinase domain. | | 分子名称: | Calmodulin-like domain protein kinase isoform 3 | | 著者 | Wernimont, A.K, Lew, J, Kozieradzki, I, Cossar, D, Wasney, G, Lin, Y.H, Hassani, A, Ali, A, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Wikstrom, M, Edwards, A.M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | 登録日 | 2008-07-24 | | 公開日 | 2008-09-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Crystal structure of the calcium-dependent kinase from toxoplasma gondii, 541.m00134, kinase domain.
To be Published
|
|
3ZDT
 
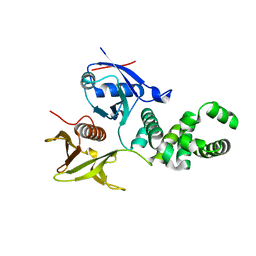 | | Crystal structure of basic patch mutant FAK FERM domain FAK31- 405 K216A, K218A, R221A, K222A | | 分子名称: | FOCAL ADHESION KINASE 1 | | 著者 | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | 登録日 | 2012-11-30 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7ZRI
 
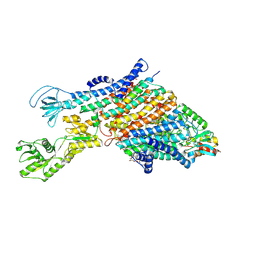 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | 分子名称: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
3S9B
 
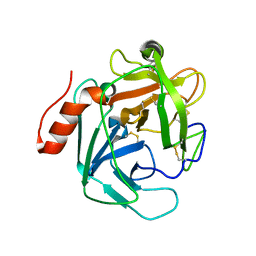 | |
5T0U
 
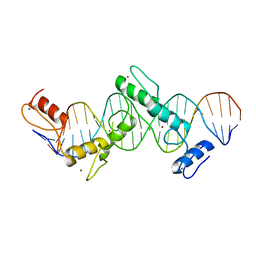 | | CTCF ZnF2-7 and DNA complex structure | | 分子名称: | DNA (5'-D(*CP*CP*TP*CP*AP*CP*TP*AP*GP*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*CP*TP*AP*GP*TP*GP*AP*GP*G)-3'), Transcriptional repressor CTCF, ... | | 著者 | Hashimoto, H, Wang, D, Cheng, X. | | 登録日 | 2016-08-16 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.199 Å) | | 主引用文献 | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
2IJ3
 
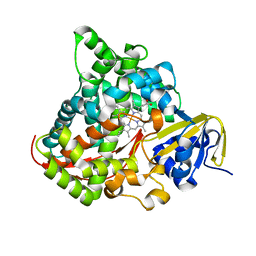 | | Structure of the A264H mutant of cytochrome P450 BM3 | | 分子名称: | Cytochrome P450 BM3, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Toogood, H.S, Leys, D. | | 登録日 | 2006-09-29 | | 公開日 | 2006-11-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and spectroscopic characterization of P450 BM3 mutants with unprecedented P450 heme iron ligand sets. New heme ligation states influence conformational equilibria in P450 BM3.
J.Biol.Chem., 282, 2007
|
|
3SI1
 
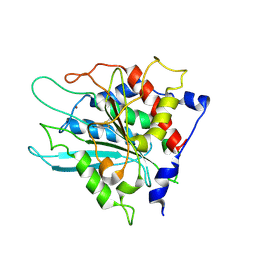 | | Structure of glycosylated murine glutaminyl cyclase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutaminyl-peptide cyclotransferase, ZINC ION | | 著者 | Dambe, T, Carrillo, D, Parthier, C, Stubbs, M.T. | | 登録日 | 2011-06-17 | | 公開日 | 2011-06-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures of Glycosylated Mammalian Glutaminyl Cyclases Reveal Conformational Variability near the Active Center.
Biochemistry, 50, 2011
|
|
3ZGE
 
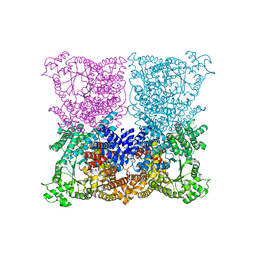 | | Greater efficiency of photosynthetic carbon fixation due to single amino acid substitution | | 分子名称: | 1,2-ETHANEDIOL, ASPARTIC ACID, C4 PHOSPHOENOLPYRUVATE CARBOXYLASE, ... | | 著者 | Paulus, J.K, Schlieper, D, Groth, G. | | 登録日 | 2012-12-17 | | 公開日 | 2013-02-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Greater Efficiency of Photosynthetic Carbon Fixation due to Single Amino Acid Substitution
Nat.Commun., 4, 2013
|
|
2IJR
 
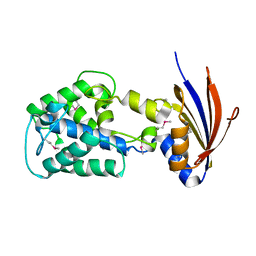 | | Crystal structure of a protein api92 from Yersinia pseudotuberculosis, Pfam DUF1281 | | 分子名称: | Hypothetical protein api92 | | 著者 | Jin, X, Min, T, Bonanno, J.B, Sauder, J.M, Wasserman, S, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2006-09-30 | | 公開日 | 2006-10-31 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of a hypothetical protein from Yersinia
pseudotuberculosis
To be Published
|
|
5T1J
 
 | | Crystal Structure of the Tbox DNA binding domain of the transcription factor T-bet | | 分子名称: | DNA, T-box transcription factor TBX21 | | 著者 | Liu, C.F, Brandt, G.S, Hoang, Q, Hwang, E.S, Naumova, N, Lazarevic, V, Dekker, J, Glimcher, L.H, Ringe, D, Petsko, G.A. | | 登録日 | 2016-08-19 | | 公開日 | 2016-10-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.947 Å) | | 主引用文献 | Crystal structure of the DNA binding domain of the transcription factor T-bet suggests simultaneous recognition of distant genome sites.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3ER5
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | 分子名称: | ENDOTHIAPEPSIN, H-189 | | 著者 | Bailey, D, Veerapandian, B, Cooper, J, Szelke, M, Blundell, T.L. | | 登録日 | 1991-01-05 | | 公開日 | 1991-04-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray-crystallographic studies of complexes of pepstatin A and a statine-containing human renin inhibitor with endothiapepsin.
Biochem.J., 289 ( Pt 2), 1993
|
|
