6XLV
 
 | |
1E3U
 
 | | MAD structure of OXA10 class D beta-lactamase | | 分子名称: | 1,2-ETHANEDIOL, BETA-LACTAMASE OXA-10, GOLD (I) CYANIDE ION, ... | | 著者 | Maveyraud, L, Golemi, D, Kotra, L.P, Tranier, S, Vakulenko, S, Mobashery, S, Samama, J.P. | | 登録日 | 2000-06-23 | | 公開日 | 2001-01-12 | | 最終更新日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Insights Into Class D Beta-Lactamases are Revealed by the Crystal Structure of the Oxa10 Enzyme from Pseudomonas Aeruginosa
Structure, 8, 2000
|
|
8UK6
 
 | |
7NK0
 
 | | Structure of the BIR1 domain of cIAP2 | | 分子名称: | Baculoviral IAP repeat-containing protein 3, ZINC ION | | 著者 | Cossu, F, Milani, M, Mastrangelo, E, Mirdita, D. | | 登録日 | 2021-02-17 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure-based identification of a new IAP-targeting compound that induces cancer cell death inducing NF-kappa B pathway.
Comput Struct Biotechnol J, 19, 2021
|
|
6Y9J
 
 | | Crystal Structure of subtype-switched Epithelial Adhesin 1 to 9 A domain (Epa1-CBL2Epa9) from Candida glabrata in complex with beta-lactose | | 分子名称: | CALCIUM ION, CHLORIDE ION, Epa1p, ... | | 著者 | Hoffmann, D, Diderrich, R, Kock, M, Friederichs, S, Reithofer, V, Essen, L.-O, Moesch, H.-U. | | 登録日 | 2020-03-09 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Functional reprogramming ofCandida glabrataepithelial adhesins: the role of conserved and variable structural motifs in ligand binding.
J.Biol.Chem., 295, 2020
|
|
6YCQ
 
 | | Crystal structure of the DNA binding domain of Arabidopsis thaliana Auxin Response Factor 1 (AtARF1) in complex with High Affinity DNA | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 21-7A, 21-7B, ... | | 著者 | Crespo, I, Weijers, D, Boer, D.R. | | 登録日 | 2020-03-18 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis .
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5IH2
 
 | | Structure, thermodynamics, and the role of conformational dynamics in the interactions between the N-terminal SH3 domain of CrkII and proline-rich motifs in cAbl | | 分子名称: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Adapter molecule crk, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Bhatt, V.S, Zeng, D, Krieger, I, Sacchettini, J.C, Cho, J.-H. | | 登録日 | 2016-02-28 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Binding Mechanism of the N-Terminal SH3 Domain of CrkII and Proline-Rich Motifs in cAbl.
Biophys.J., 110, 2016
|
|
5IH7
 
 | |
5IEY
 
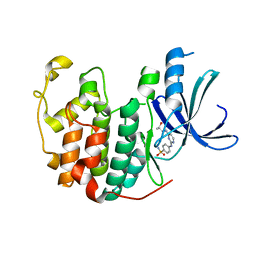 | | Crystal structure of a CDK inhibitor bound to CDK2 | | 分子名称: | 4-[(4-{[(2R,3R)-3-hydroxybutan-2-yl]amino}pyrimidin-2-yl)amino]benzene-1-sulfonamide, Cyclin-dependent kinase 2 | | 著者 | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | 登録日 | 2016-02-25 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
8TXA
 
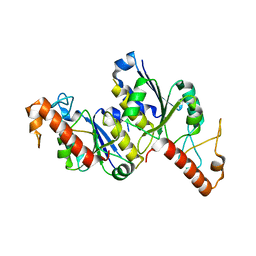 | | Apo Structure of (N1G37) tRNA Methyltransferase from Mycobacterium marinum | | 分子名称: | tRNA (guanine-N(1)-)-methyltransferase | | 著者 | Balsamo, A, Bruno, C, Edele, C, Fabian, T, Jannotta, R, Lee, R, Schryver, D, Warsaw, J, Warsaw, L, Stojanoff, V, Battaile, K, Perez, A, Bolen, R. | | 登録日 | 2023-08-23 | | 公開日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.591 Å) | | 主引用文献 | Apo Structure of (N1G37) tRNA Methyltransferase from Mycobacterium marinum
To Be Published
|
|
5IFI
 
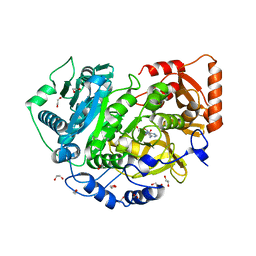 | | CRYSTAL STRUCTURE OF ACETYL-COA SYNTHETASE IN COMPLEX WITH ADENOSINE-5'-PROPYLPHOSPHATE FROM CRYPTOCOCCUS NEOFORMANS H99 | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-MONOPHOSPHATE-PROPYL ESTER, Acetyl-coenzyme A synthetase, ... | | 著者 | Seattle Structural Genomics Center for Infectious Disease (SSGCID), SSGCID, Fox III, D, Edwards, T.E, Lorimer, D.D, Mutz, M.W. | | 登録日 | 2016-02-26 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of Acetyl-CoA Synthetase in complex with Adenosine-5'-propylphosphate from Cryptococcus neoformans H99
To Be Published
|
|
6YKI
 
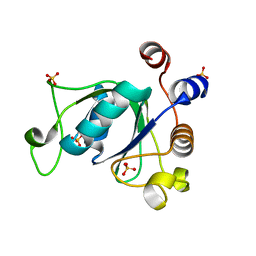 | | Crystal structure of YTHDC1 with compound DHU_DC1_092 | | 分子名称: | SULFATE ION, YTHDC1, ~{N}-ethyl-2-[(2~{S},5~{R})-5-methyl-2-phenyl-morpholin-4-yl]ethanamine | | 著者 | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | 登録日 | 2020-04-06 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
1T2X
 
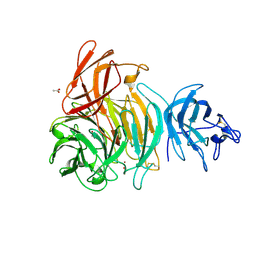 | | Glactose oxidase C383S mutant identified by directed evolution | | 分子名称: | ACETATE ION, COPPER (II) ION, Galactose Oxidase, ... | | 著者 | Wilkinson, D, Akumanyi, N, Hurtado-Guerrero, R, Dawkes, H, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | 登録日 | 2004-04-23 | | 公開日 | 2004-05-18 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and kinetic studies of a series of mutants of galactose oxidase identified by directed evolution.
Protein Eng.Des.Sel., 17, 2004
|
|
6YLT
 
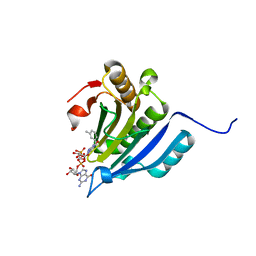 | | Translation initiation factor 4E in complex with 3-MeBn7GpppG mRNA 5' cap analog | | 分子名称: | Eukaryotic translation initiation factor 4E, [[(2~{R},3~{S},4~{R},5~{R})-5-[2-azanyl-7-[(3-methylphenyl)methyl]-6-oxidanylidene-1~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] hydrogen phosphate | | 著者 | Kubacka, D, Wojcik, R, Baranowski, M.R, Kowalska, J, Jemielity, J. | | 登録日 | 2020-04-07 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Novel N7-Arylmethyl Substituted Dinucleotide mRNA 5' cap Analogs: Synthesis and Evaluation as Modulators of Translation.
Pharmaceutics, 13, 2021
|
|
5II8
 
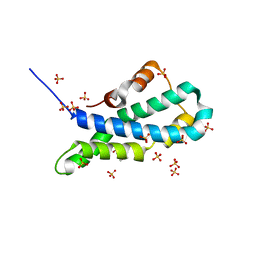 | | Orthorhombic crystal structure of red abalone lysin at 0.99 A resolution | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Egg-lysin, SULFATE ION | | 著者 | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, De Sanctis, D, Jovine, L. | | 登録日 | 2016-03-01 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (0.99 Å) | | 主引用文献 | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5IIZ
 
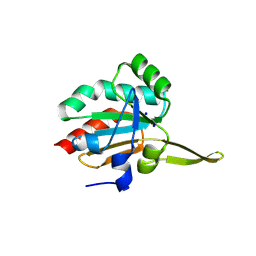 | | Xanthomonas campestris Peroxiredoxin Q - Structure F0 | | 分子名称: | Bacterioferritin comigratory protein, SODIUM ION | | 著者 | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | 登録日 | 2016-03-01 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IMC
 
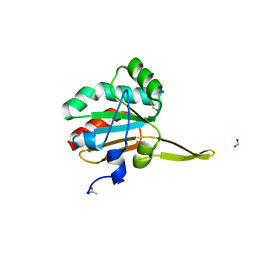 | | Xanthomonas campestris Peroxiredoxin Q - Structure F3 | | 分子名称: | Bacterioferritin comigratory protein, FORMIC ACID, SODIUM ION | | 著者 | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | 登録日 | 2016-03-06 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
8UTA
 
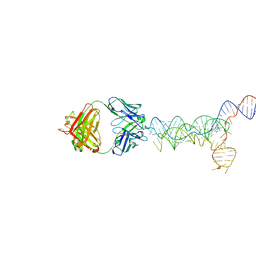 | | yjdF riboswitch from R. gauvreauii in complex with proflavine bound to Fab BL3-6 S97N | | 分子名称: | Fab BL3-6 S97N heavy chain, Fab BL3-6 S97N light chain, MAGNESIUM ION, ... | | 著者 | Krochmal, D, Lewicka, A, Piccirilli, J.A. | | 登録日 | 2023-10-30 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structural basis for promiscuity in ligand recognition by yjdF riboswitch.
Cell Discov, 10, 2024
|
|
6Y2F
 
 | | Crystal structure (monoclinic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | 著者 | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | 登録日 | 2020-02-15 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
5IO0
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F9 | | 分子名称: | Bacterioferritin comigratory protein, SODIUM ION | | 著者 | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | 登録日 | 2016-03-08 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
8UKU
 
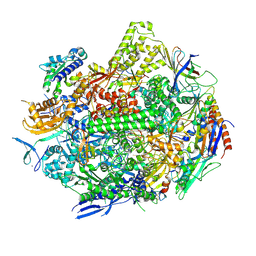 | | RNA polymerase II elongation complex with Fapy-dG lesion with CMP added | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | 著者 | Hou, P, Oh, J, Wang, D. | | 登録日 | 2023-10-15 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Molecular Mechanism of RNA Polymerase II Transcriptional Mutagenesis by the Epimerizable DNA Lesion, Fapy·dG.
J.Am.Chem.Soc., 146, 2024
|
|
5IOW
 
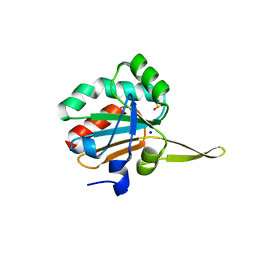 | | Xanthomonas campestris Peroxiredoxin Q - Structure FFcumene (Hyperoxidized by cumene hydroperoxide) | | 分子名称: | Bacterioferritin comigratory protein, SODIUM ION | | 著者 | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | 登録日 | 2016-03-09 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
8U4K
 
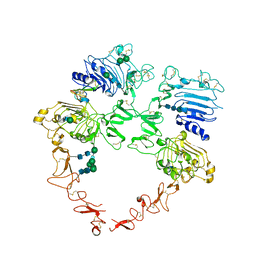 | | Structure of the HER2/HER4/BTC Heterodimer Extracellular Domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Betacellulin, ... | | 著者 | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | 登録日 | 2023-09-10 | | 公開日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.27 Å) | | 主引用文献 | Structure of the HER2/HER4/BTC Heterodimer Extracellular Domain
To Be Published
|
|
8U4I
 
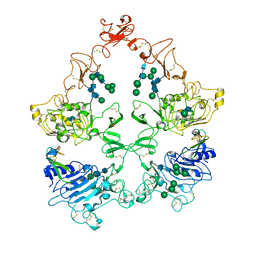 | | Structure of the HER4/NRG1b Homodimer Extracellular Domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | 著者 | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | 登録日 | 2023-09-10 | | 公開日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | Structure of the HER2/HER4/NRG1b Heterodimer Extracellular Domain
To Be Published
|
|
5IQL
 
 | |
