7GD1
 
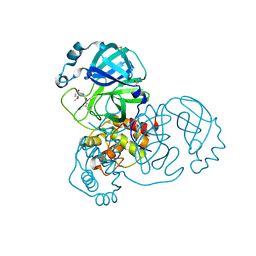 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BAR-COM-0f94fc3d-58 (Mpro-x10856) | | 分子名称: | (2R)-2-amino-2-(5-bromo-2-methoxyphenyl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
3X1O
 
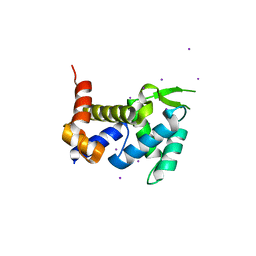 | | Crystal structure of the ROQ domain of human Roquin | | 分子名称: | IODIDE ION, Roquin-1 | | 著者 | Ose, T, Verma, A, Cockburn, J.B, Berrow, N.S, Alderton, D, Stuart, D, Owens, R.J, Jones, E.Y. | | 登録日 | 2014-11-26 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | Roquin binds microRNA-146a and Argonaute2 to regulate microRNA homeostasis
Nat Commun, 6, 2015
|
|
7BN2
 
 | |
4IXQ
 
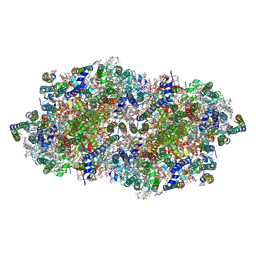 | | RT fs X-ray diffraction of Photosystem II, dark state | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Kern, J, Alonso-Mori, R, Tran, R, Hattne, J, Gildea, R.J, Echols, N, Gloeckner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Koroidov, S, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, Schafer, D.W, Messerschmidt, M, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Grosse-Kunstleve, R.W, Zwart, P.H, White, W.E, Glatzel, P, Adams, P.D, Bogan, M.J, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Sauter, N.K, Yachandra, V.K, Bergmann, U, Yano, J. | | 登録日 | 2013-01-27 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (5.7 Å) | | 主引用文献 | Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature.
Science, 340, 2013
|
|
6TIH
 
 | |
3NQ8
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution R4 8/5A | | 分子名称: | BENZAMIDINE, NITRATE ION, deoxyribose phosphate aldolase | | 著者 | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2010-06-29 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
6TM6
 
 | | MUC2 CysD1 domain | | 分子名称: | CALCIUM ION, Mucin-2 | | 著者 | Khmelnitsky, L, Fass, D. | | 登録日 | 2019-12-03 | | 公開日 | 2020-02-19 | | 最終更新日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Assembly Mechanism of Mucin and von Willebrand Factor Polymers.
Cell, 183, 2020
|
|
6T8V
 
 | | Complement factor B in complex with (S)-5,7-Dimethyl-4-((2-phenylpiperidin-1-yl)methyl)-1H-indole | | 分子名称: | 4-[(2~{S})-1-[(5,7-dimethyl-1~{H}-indol-4-yl)methyl]piperidin-2-yl]benzoic acid, Complement factor B, SULFATE ION, ... | | 著者 | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | 登録日 | 2019-10-25 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
3ZUF
 
 | |
3ZUJ
 
 | |
1KRX
 
 | | SOLUTION STRUCTURE OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN: MODEL STRUCTURES INCORPORATING ACTIVE SITE CONTACTS | | 分子名称: | BERYLLIUM TRIFLUORIDE ION, NITROGEN REGULATION PROTEIN NR(I) | | 著者 | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | 登録日 | 2002-01-10 | | 公開日 | 2003-08-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
4RRS
 
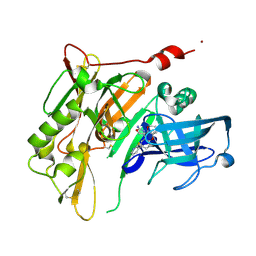 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | 分子名称: | (4R,4a'R,10a'S)-8'-(2-fluoropyridin-3-yl)-4a'-methyl-3',4',4a',10a'-tetrahydro-2'H-spiro[1,3-oxazole-4,10'-pyrano[3,2-b]chromen]-2-amine, Beta-secretase 1, NICKEL (II) ION | | 著者 | Thomas, A.A, Hunt, K.W, Newhouse, B, Watts, R.J, Liu, X, Vigers, G.P.A, Smith, D, Rhodes, S.P, Brown, K.D, Otten, J.N, Burkard, M, Cox, A.A, Geck Do, M.K, Dutcher, D, Rana, S, DeLisle, R.K, Regal, K, Wright, A.D, Groneberg, R, Liao, J, Scearce-Levie, K, Siu, M, Purkey, H.E, Lyssikatos, J.P. | | 登録日 | 2014-11-06 | | 公開日 | 2014-12-03 | | 最終更新日 | 2014-12-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
4RO5
 
 | | Crystal structure of the SAT domain from the non-reducing fungal polyketide synthase CazM | | 分子名称: | GLYCEROL, SAT domain from CazM | | 著者 | Winter, J.M, Cascio, D, Sawaya, M.R, Tang, Y. | | 登録日 | 2014-10-27 | | 公開日 | 2015-09-09 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis.
J.Am.Chem.Soc., 137, 2015
|
|
4RRO
 
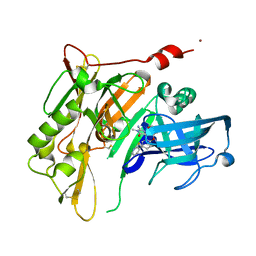 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | 分子名称: | (4S,4a'R,10a'S)-2-amino-8'-(2-fluoropyridin-3-yl)-1,4a'-dimethyl-3',4',4a',10a'-tetrahydro-2'H-spiro[imidazole-4,10'-pyrano[3,2-b]chromen]-5(1H)-one, Beta-secretase 1, NICKEL (II) ION | | 著者 | Thomas, A.A, Hunt, K.W, Newhouse, B, Watts, R.J, Liu, X, Vigers, G.P.A, Smith, D, Rhodes, S.P, Brown, K.D, Otten, J.N, Burkard, M, Cox, A.A, Geck Do, M.K, Dutcher, D, Rana, S, DeLisle, R.K, Regal, K, Wright, A.D, Groneberg, R, Liao, J, Scearce-Levie, K, Siu, M, Purkey, H.E, Lyssikatos, J.P. | | 登録日 | 2014-11-06 | | 公開日 | 2014-12-03 | | 最終更新日 | 2014-12-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
1KRW
 
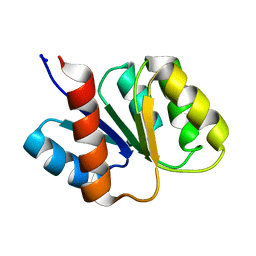 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN | | 分子名称: | NITROGEN REGULATION PROTEIN NR(I) | | 著者 | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | 登録日 | 2002-01-10 | | 公開日 | 2003-08-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
2P4E
 
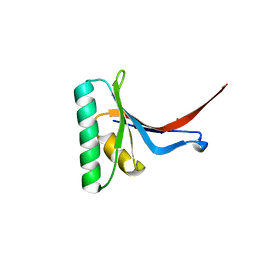 | | Crystal Structure of PCSK9 | | 分子名称: | MERCURY (II) ION, Proprotein convertase subtilisin/kexin type 9 | | 著者 | Cunningham, D, Danley, D.E, Geoghegan, F.K, Griffor, M.C, Hawkins, J.L, Qiu, X. | | 登録日 | 2007-03-12 | | 公開日 | 2007-04-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia.
Nat.Struct.Mol.Biol., 14, 2007
|
|
3UMI
 
 | | X-ray structure of the E2 domain of the human amyloid precursor protein (APP) in complex with zinc | | 分子名称: | ACETATE ION, Amyloid beta A4 protein, CADMIUM ION, ... | | 著者 | Dahms, S.O, Konnig, I, Roeser, D, Guhrs, K.H, Than, M.E. | | 登録日 | 2011-11-13 | | 公開日 | 2012-01-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Metal Binding Dictates Conformation and Function of the Amyloid Precursor Protein (APP) E2 Domain.
J.Mol.Biol., 416, 2012
|
|
2KWI
 
 | | RalB-RLIP76 (RalBP1) complex | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RalA-binding protein 1, ... | | 著者 | Fenwick, R.B, Campbell, L.J, Rajasekar, K, Prasannan, S, Nietlispach, D, Camonis, J, Owen, D, Mott, H.R. | | 登録日 | 2010-04-12 | | 公開日 | 2010-09-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The RalB-RLIP76 complex reveals a novel mode of ral-effector interaction
Structure, 18, 2010
|
|
2KWH
 
 | | Ral binding domain of RLIP76 (RalBP1) | | 分子名称: | RalA-binding protein 1 | | 著者 | Fenwick, R.B, Campbell, L.J, Rajasekar, K, Prasannan, S, Nietlispach, D, Camonis, J, Owen, D, Mott, H.R. | | 登録日 | 2010-04-12 | | 公開日 | 2010-09-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The RalB-RLIP76 complex reveals a novel mode of ral-effector interaction
Structure, 18, 2010
|
|
7CYV
 
 | | Crystal structure of FD20, a neutralizing single-chain variable fragment (scFv) in complex with SARS-CoV-2 Spike receptor-binding domain (RBD) | | 分子名称: | Spike protein S1, The heavy chain variable region of the scFv FD20,The light chain variable region of the scFv FD20, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Li, Y, Li, T, Lai, Y, Cai, H, Yao, H, Li, D. | | 登録日 | 2020-09-04 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | Uncovering a conserved vulnerability site in SARS-CoV-2 by a human antibody.
Embo Mol Med, 13, 2021
|
|
1I8F
 
 | | THE CRYSTAL STRUCTURE OF A HEPTAMERIC ARCHAEAL SM PROTEIN: IMPLICATIONS FOR THE EUKARYOTIC SNRNP CORE | | 分子名称: | GLYCEROL, PUTATIVE SNRNP SM-LIKE PROTEIN | | 著者 | Mura, C, Cascio, D, Sawaya, M.R, Eisenberg, D. | | 登録日 | 2001-03-14 | | 公開日 | 2001-05-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The crystal structure of a heptameric archaeal Sm protein: Implications for the eukaryotic snRNP core.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7F5G
 
 | | The crystal structure of RBD-Nanobody complex, DL4 (SA4) | | 分子名称: | ACETATE ION, GLYCEROL, Nanobody DL4, ... | | 著者 | Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | 登録日 | 2021-06-22 | | 公開日 | 2022-05-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Isolation, characterization, and structure-based engineering of a neutralizing nanobody against SARS-CoV-2.
Int.J.Biol.Macromol., 209, 2022
|
|
2PEA
 
 | |
1LIT
 
 | | HUMAN LITHOSTATHINE | | 分子名称: | LITHOSTATHINE | | 著者 | Bertrand, J.A, Pignol, D, Bernard, J.-P, Verdier, J.-M, Dagorn, J.-C, Fontacilla-Camps, J.C. | | 登録日 | 1996-01-17 | | 公開日 | 1997-01-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structure of human lithostathine, the pancreatic inhibitor of stone formation.
EMBO J., 15, 1996
|
|
4B00
 
 | | Design and Synthesis of BACE1 Inhibitors with In Vivo Brain Reduction of beta-Amyloid Peptides (COMPOUND (R)-41) | | 分子名称: | 5-{(1R)-3-amino-4-fluoro-1-[3-(5-prop-1-yn-1-ylpyridin-3-yl)phenyl]-1H-isoindol-1-yl}-1-ethyl-3-methylpyridin-2(1H)-one, ACETATE ION, BETA-SECRETASE 1 | | 著者 | Swahn, B.M, Kolmodin, K, Karlstrom, S, von Berg, S, Soderman, P, Holenz, J, Berg, S, Lindstrom, J, Sundstrom, M, Turek, D, Kihlstrom, J, Slivo, C, Andersson, L, Pyring, D, Ohberg, L, Kers, A, Bogar, K, Bergh, M, Olsson, L.L, Janson, J, Eketjall, S, Georgievska, B, Jeppsson, F, Falting, J. | | 登録日 | 2012-06-27 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Design and synthesis of beta-site amyloid precursor protein cleaving enzyme (BACE1) inhibitors with in vivo brain reduction of beta-amyloid peptides.
J. Med. Chem., 55, 2012
|
|
