6CN3
 
 | |
5D3J
 
 | | First bromodomain of BRD4 bound to inhibitor XD33 | | 分子名称: | 4-acetyl-N-[3-(diethylsulfamoyl)phenyl]-3-ethyl-5-methyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | 著者 | Wohlwend, D, Huegle, M. | | 登録日 | 2015-08-06 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
5D3R
 
 | | First bromodomain of BRD4 bound to inhibitor XD42 | | 分子名称: | 4-acetyl-N-[3-(azepan-1-ylsulfonyl)phenyl]-5-methyl-3-propyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | 著者 | Wohlwend, D, Huegle, M. | | 登録日 | 2015-08-06 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
6CGO
 
 | | Molecular basis for condensation domain-mediated chain release from the enacyloxin polyketide synthase | | 分子名称: | Condensation domain protein, PHOSPHATE ION | | 著者 | Valentic, T.R, Tsai, S.C, Challis, G.L, Lewandowski, J.R, Kosol, S, Gallo, A, Griffiths, D, Masschelein, J.L, Jenner, M, De los Santos, E. | | 登録日 | 2018-02-20 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for chain release from the enacyloxin polyketide synthase.
Nat.Chem., 11, 2019
|
|
1PUL
 
 | | Solution structure for the 21KDa caenorhabditis elegans protein CE32E8.3. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET WR33 | | 分子名称: | Hypothetical protein C32E8.3 in chromosome I | | 著者 | Tejero, R, Aramini, J.M, Swapna, G.V.T, Monleon, D, Chiang, Y, Macapagal, D, Gunsalus, K.C, Kim, S, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2003-06-25 | | 公開日 | 2005-06-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Backbone 1H, 15N and 13C assignments for the 21 kDa Caenorhabditis elegans homologue of "brain-specific" protein.
J.Biomol.Nmr, 28, 2004
|
|
5AN7
 
 | | Structure of the engineered retro-aldolase RA95.5-8F with a bound 1,3-diketone inhibitor | | 分子名称: | (2E)-1-(6-methoxynaphthalen-2-yl)but-2-en-1-one, PHOSPHATE ION, RA95.5-8F | | 著者 | Obexer, R, Mittl, P.R.E, Hilvert, D. | | 登録日 | 2015-09-04 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase.
Nat Chem, 9, 2017
|
|
3C5R
 
 | |
1PZ2
 
 | | Crystal structure of a transient covalent reaction intermediate of a family 51 alpha-L-arabinofuranosidase | | 分子名称: | Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose | | 著者 | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Baasov, T, Shoham, Y, Schomburg, D. | | 登録日 | 2003-07-09 | | 公開日 | 2003-10-07 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
1Q90
 
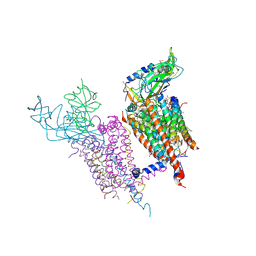 | | Structure of the cytochrome b6f (plastohydroquinone : plastocyanin oxidoreductase) from Chlamydomonas reinhardtii | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 8-HYDROXY-5,7-DIMETHOXY-3-METHYL-2-TRIDECYL-4H-CHROMEN-4-ONE, ... | | 著者 | Stroebel, D, Choquet, Y, Popot, J.-L, Picot, D. | | 登録日 | 2003-08-22 | | 公開日 | 2003-12-09 | | 最終更新日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | An Atypical Haem in the Cytochrome B6F Complex
Nature, 426, 2003
|
|
4ZX2
 
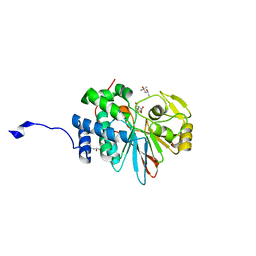 | | Co-crystal structures of PP5 in complex with 5-methyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid | | 分子名称: | (1S,2R,3S,4R,5S)-5-methyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | 著者 | Chattopadhyay, D, Swingle, M.R, Salter, E.A, Wierzbicki, A, Honkanen, R.E. | | 登録日 | 2015-05-19 | | 公開日 | 2016-04-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | Crystal structures and mutagenesis of PPP-family ser/thr protein phosphatases elucidate the selectivity of cantharidin and novel norcantharidin-based inhibitors of PP5C.
Biochem. Pharmacol., 109, 2016
|
|
1NNO
 
 | | CONFORMATIONAL CHANGES OCCURRING UPON NO BINDING IN NITRITE REDUCTASE FROM PSEUDOMONAS AERUGINOSA | | 分子名称: | HEME C, HEME D, NITRIC OXIDE, ... | | 著者 | Nurizzo, D, Tegoni, M, Cambillau, C. | | 登録日 | 1998-07-20 | | 公開日 | 1999-04-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Conformational changes occurring upon reduction and NO binding in nitrite reductase from Pseudomonas aeruginosa.
Biochemistry, 37, 1998
|
|
4QC9
 
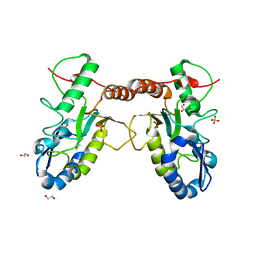 | | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant 3GD4 | | 分子名称: | 1,2-ETHANEDIOL, SULFATE ION, Uracil-DNA glycosylase | | 著者 | Sartmatova, D, Nash, T, Schormann, N, Nuth, M, Ricciardi, R, Banerjee, S, Chattopadhyay, D. | | 登録日 | 2014-05-09 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.259 Å) | | 主引用文献 | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant 3GD4
To be Published
|
|
1QMA
 
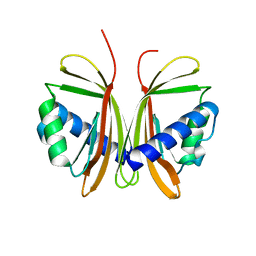 | | Nuclear Transport Factor 2 (NTF2) W7A mutant | | 分子名称: | NUCLEAR TRANSPORT FACTOR 2 | | 著者 | Bayliss, R, Ribbeck, K, Akin, D, Kent, H.M, Feldherr, C.M, Gorlich, D, Stewart, M.J. | | 登録日 | 1999-09-23 | | 公開日 | 2000-02-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Interaction Betweeen Ntf2 and Xfxfg-Containing Nucleoporins is Required to Mediate Nuclear Import of Ran-Gdp
J.Mol.Biol., 293, 1999
|
|
5D3L
 
 | | First bromodomain of BRD4 bound to inhibitor XD35 | | 分子名称: | 4-acetyl-N-[5-(diethylsulfamoyl)-2-hydroxy-4-methylphenyl]-3-ethyl-5-methyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | 著者 | Wohlwend, D, Huegle, M. | | 登録日 | 2015-08-06 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
5D3P
 
 | | First bromodomain of BRD4 bound to inhibitor XD41 | | 分子名称: | 1-[(4-acetyl-3-ethyl-5-methyl-1H-pyrrol-2-yl)carbonyl]-N-methyl-1H-indole-6-sulfonamide, Bromodomain-containing protein 4, NICKEL (II) ION | | 著者 | Wohlwend, D, Huegle, M. | | 登録日 | 2015-08-06 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
1PK6
 
 | | Globular Head of the Complement System Protein C1q | | 分子名称: | CALCIUM ION, Complement C1q subcomponent, A chain precursor, ... | | 著者 | Gaboriaud, C, Juanhuix, J, Gruez, A, Lacroix, M, Darnault, C, Pignol, D, Verger, D, Fontecilla-Camps, J.C, Arlaud, G.J. | | 登録日 | 2003-06-05 | | 公開日 | 2003-10-21 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties.
J.Biol.Chem., 278, 2003
|
|
5D26
 
 | | First bromodomain of BRD4 bound to inhibitor XD28 | | 分子名称: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | 著者 | Wohlwend, D, Huegle, M. | | 登録日 | 2015-08-05 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
5D25
 
 | | First bromodomain of BRD4 bound to inhibitor XD27 | | 分子名称: | (R,R)-2,3-BUTANEDIOL, 4-acetyl-N-[5-(diethylsulfamoyl)-2-hydroxyphenyl]-3,5-dimethyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4, ... | | 著者 | Wohlwend, D, Huegle, M, Gerhardt, S. | | 登録日 | 2015-08-05 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
1QW9
 
 | | Crystal structure of a family 51 alpha-L-arabinofuranosidase in complex with 4-nitrophenyl-Ara | | 分子名称: | 4-nitrophenyl alpha-L-arabinofuranoside, Alpha-L-arabinofuranosidase | | 著者 | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Bassov, T, Shoham, Y, Schomburg, D. | | 登録日 | 2003-09-01 | | 公開日 | 2003-10-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
3AH7
 
 | | Crystal structure of the ISC-like [2Fe-2S] ferredoxin (FdxB) from Pseudomonas putida JCM 20004 | | 分子名称: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, SODIUM ION, ... | | 著者 | Kumasaka, T, Shimizu, N, Ohmori, D, Iwasaki, T. | | 登録日 | 2010-04-20 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the ISC-like [2Fe-2S] ferredoxin (FdxB) from Pseudomonas putida JCM 20004
To be Published
|
|
1QW8
 
 | | Crystal structure of a family 51 alpha-L-arabinofuranosidase in complex with Ara-alpha(1,3)-Xyl | | 分子名称: | Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose | | 著者 | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Bassov, T, Shoham, Y, Schomburg, D. | | 登録日 | 2003-09-01 | | 公開日 | 2003-10-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
7FSE
 
 | | Crystal Structure of T. maritima reverse gyrase with a minimal latch | | 分子名称: | CHLORIDE ION, DODECAETHYLENE GLYCOL, Reverse gyrase, ... | | 著者 | Rasche, R, Kummel, D, Rudolph, M.G, Klostermeier, D. | | 登録日 | 2023-01-04 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structure of reverse gyrase with a minimal latch that supports ATP-dependent positive supercoiling without specific interactions with the topoisomerase domain.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7FSF
 
 | | CRYSTAL STRUCTURE OF T. MARITIMA REVERSE GYRASE ACTIVE SITE VARIANT Y851F | | 分子名称: | Reverse gyrase, ZINC ION | | 著者 | Rasche, R, Kummel, D, Rudolph, M.G, Klostermeier, D. | | 登録日 | 2023-01-04 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | Structure of reverse gyrase with a minimal latch that supports ATP-dependent positive supercoiling without specific interactions with the topoisomerase domain.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
3CLR
 
 | | Crystal structure of the R236A ETF mutant from M. methylotrophus | | 分子名称: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | 著者 | Katona, G, Leys, D. | | 登録日 | 2008-03-20 | | 公開日 | 2008-04-08 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
3CW9
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase in the Thioester-forming Conformation, bound to 4-chlorophenacyl-CoA | | 分子名称: | 1,2-ETHANEDIOL, 4-Chlorophenacyl-coenzyme A, 4-chlorobenzoyl CoA ligase, ... | | 著者 | Reger, A.S, Cao, J, Wu, R, Dunaway-Mariano, D, Gulick, A.M. | | 登録日 | 2008-04-21 | | 公開日 | 2008-09-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase.
Biochemistry, 47, 2008
|
|
