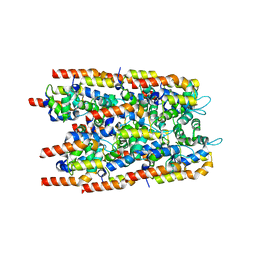
6I3N
 
 | |
6IB1
 
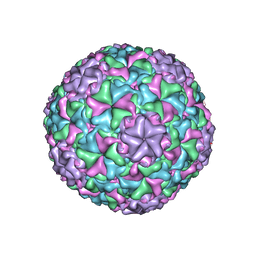 | | Icosahedrally averaged capsid of empty particle of bacteriophage P68 | | 分子名称: | Major head protein, Uncharacterized protein | | 著者 | Hrebik, D, Skubnik, K, Fuzik, T, Plevka, P. | | 登録日 | 2018-11-28 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure and genome ejection mechanism ofStaphylococcus aureusphage P68.
Sci Adv, 5, 2019
|
|
4ZA8
 
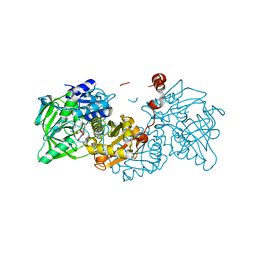 | | Crystal structure of A niger Fdc1 in complex with penta-fluorocinnamic acid | | 分子名称: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 1-deoxy-5-O-phosphono-1-[(10aR)-2,2,3,4-tetramethyl-8,10-dioxo-1,2,8,9,10,10a-hexahydro-6H-indeno[1,7-ef]pyrimido[4,5-b][1,4]diazepin-6-yl]-D-ribitol, MANGANESE (II) ION, ... | | 著者 | Payne, K.A.P, Leys, D. | | 登録日 | 2015-04-13 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
4ZAL
 
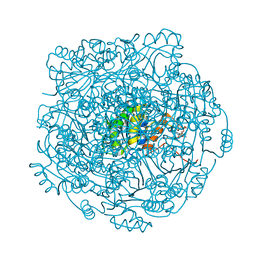 | | Structure of UbiX E49Q mutant in complex with reduced FMN and dimethylallyl monophosphate | | 分子名称: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Dimethylallyl monophosphate, THIOCYANATE ION, ... | | 著者 | White, M.D, Leys, D. | | 登録日 | 2015-04-13 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
6ZK1
 
 | | Plant nucleoside hydrolase - ZmNRh2b enzyme | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Morera, S, Vigouroux, A, Kopecny, D. | | 登録日 | 2020-06-29 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK2
 
 | | Plant nucleoside hydrolase - ZmNRh2b in complex with forodesine | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, ... | | 著者 | Morera, S, Vigouroux, A, Kopecny, D. | | 登録日 | 2020-06-29 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK3
 
 | | Plant nucleoside hydrolase - ZmNRh2b in complex with ribose | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Morera, S, Vigouroux, A, Kopecny, D. | | 登録日 | 2020-06-29 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK4
 
 | | Plant nucleoside hydrolase - ZmNRh2b with a bound adenine | | 分子名称: | 1,2-ETHANEDIOL, ADENINE, CALCIUM ION, ... | | 著者 | Morera, S, Vigouroux, A, Kopecny, D. | | 登録日 | 2020-06-29 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
6ZK5
 
 | | Plant nucleoside hydrolase - ZmNRh3 enzyme in complex with forodesine | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, ... | | 著者 | Morera, S, Vigouroux, A, Kopecny, D. | | 登録日 | 2020-06-29 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Plant nucleoside N-ribohydrolases: riboside binding and role in nitrogen storage mobilization.
Plant J., 2023
|
|
4ZAB
 
 | | Structure of A. niger Fdc1 in complex with alpha-fluoro cinnamic acid | | 分子名称: | (2Z)-2-fluoro-3-phenylprop-2-enoic acid, 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 1-deoxy-5-O-phosphono-1-[(10aR)-2,2,3,4-tetramethyl-8,10-dioxo-1,2,8,9,10,10a-hexahydro-6H-indeno[1,7-ef]pyrimido[4,5-b][1,4]diazepin-6-yl]-D-ribitol, ... | | 著者 | Payne, K.A.P, Leys, D. | | 登録日 | 2015-04-13 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
4ZAW
 
 | | Structure of UbiX in complex with reduced prenylated FMN | | 分子名称: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, PHOSPHATE ION, Probable aromatic acid decarboxylase, ... | | 著者 | White, M.D, Leys, D. | | 登録日 | 2015-04-14 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
4WTD
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS AND DELTA8 BETA HAIRPIN LOOP DELETION IN COMPLEX WITH ADP, MN2+ AND SYMMETRICAL PRIMER TEMPLATE 5'-AUAAAUUU | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MANGANESE (II) ION, ... | | 著者 | Edwards, T.E, Fox III, D, Appleby, T.C. | | 登録日 | 2014-10-29 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTM
 
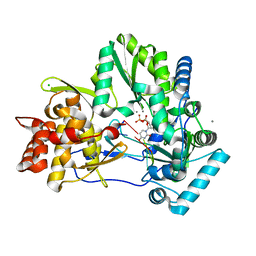 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-UAGG, RNA PRIMER 5'-PCC, MN2+, AND UDP | | 分子名称: | CHLORIDE ION, MANGANESE (II) ION, RNA PRIMER CC, ... | | 著者 | Edwards, T.E, Appleby, T.C, Fox III, D. | | 登録日 | 2014-10-30 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4ZAC
 
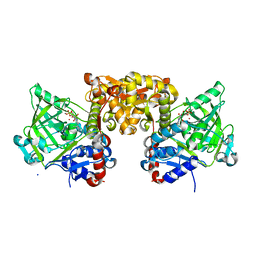 | | Structure of S. cerevisiae Fdc1 with the prenylated-flavin cofactor in the iminium form. | | 分子名称: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | 著者 | White, M.D, Leys, D. | | 登録日 | 2015-04-13 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
5ZU1
 
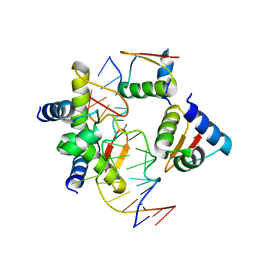 | | Crystal Structure of BZ junction in diverse sequence | | 分子名称: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*AP*GP*GP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*CP*TP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | 著者 | Kim, K.K, Kim, D. | | 登録日 | 2018-05-05 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.009 Å) | | 主引用文献 | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
6IAT
 
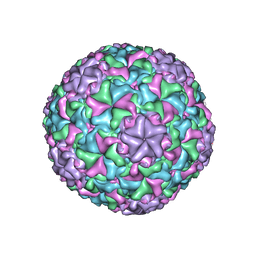 | | Icosahedrally averaged capsid of bacteriophage P68 | | 分子名称: | Arstotzka protein, Major head protein | | 著者 | Hrebik, D, Skubnik, K, Fuzik, T, Plevka, P. | | 登録日 | 2018-11-27 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure and genome ejection mechanism ofStaphylococcus aureusphage P68.
Sci Adv, 5, 2019
|
|
8CHW
 
 | |
8CHT
 
 | |
8CHV
 
 | |
6ICE
 
 | | Crystal structure of Hamster MIF | | 分子名称: | Macrophage migration inhibitory factor | | 著者 | Sundaram, R, Vasudevan, D. | | 登録日 | 2018-09-05 | | 公開日 | 2019-09-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Macrophage migration inhibitory factor of Syrian golden hamster shares structural and functional similarity with human counterpart and promotes pancreatic cancer.
Sci Rep, 9, 2019
|
|
8CJ9
 
 | | Crystal structure of maize CKO/CKX8 in complex with urea-derived inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]benzamide | | 分子名称: | 1,2-ETHANEDIOL, 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]benzamide, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Kopecny, D, Briozzo, P, Morera, S. | | 登録日 | 2023-02-12 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 2024
|
|
8CHU
 
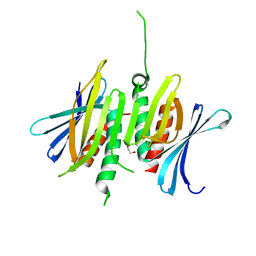 | |
8CHF
 
 | | cryo-EM Structure of Craf:14-3-3:Mek1 | | 分子名称: | 14-3-3 protein zeta isoform X1, 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, Dual specificity mitogen-activated protein kinase kinase 1, ... | | 著者 | Dedden, D, Ulrich, G. | | 登録日 | 2023-02-07 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.25 Å) | | 主引用文献 | Cryo-EM Structures of CRAF 2 /14-3-3 2 and CRAF 2 /14-3-3 2 /MEK1 2 Complexes.
J.Mol.Biol., 436, 2024
|
|
4WZB
 
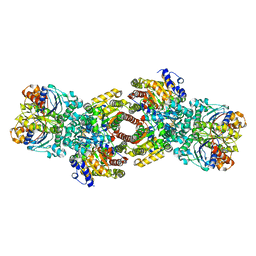 | | Crystal Structure of MgAMPPCP-bound Av2-Av1 complex | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (II) ION, FE(8)-S(7) CLUSTER, ... | | 著者 | Tezcan, F.A, Kaiser, J.T, Mustafi, D, Walton, M.Y, Howard, J.B, Rees, D.C. | | 登録日 | 2014-11-19 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Nitrogenase complexes: multiple docking sites for a nucleotide switch protein.
Science, 309, 2005
|
|
6I83
 
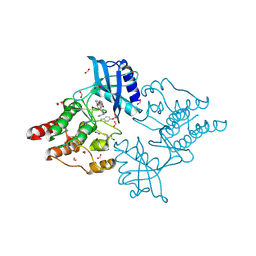 | | Crystal structure of phosphorylated RET V804M tyrosine kinase domain complexed with PDD00018366 | | 分子名称: | 4-[5-(pyridin-3-ylmethylamino)pyrazolo[1,5-a]pyrimidin-3-yl]benzamide, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | 著者 | Burschowsky, D, Seewooruthun, C, Bayliss, R, Carr, M.D, Echalier, A, Jordan, A.M. | | 登録日 | 2018-11-19 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Discovery and Optimization of wt-RET/KDR-Selective Inhibitors of RETV804MKinase.
Acs Med.Chem.Lett., 11, 2020
|
|
