3TF3
 
 | |
3TH7
 
 | |
3THJ
 
 | |
2WVI
 
 | | Crystal Structure of the N-terminal Domain of BubR1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MITOTIC CHECKPOINT SERINE/THREONINE-PROTEIN KINASE BUB1 BETA, ... | | Authors: | D'Arcy, S, Davies, O.R, Blundell, T.L, Bolanos-Garcia, V.M. | | Deposit date: | 2009-10-16 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Defining the Molecular Basis of Bubr1 Kinetochore Interactions and Anaphase-Promoting Complex/Cyclosome (Apc/C)-Cdc20 Inhibition
J.Biol.Chem., 285, 2010
|
|
3THH
 
 | |
1F9A
 
 | | CRYSTAL STRUCTURE ANALYSIS OF NMN ADENYLYLTRANSFERASE FROM METHANOCOCCUS JANNASCHII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HYPOTHETICAL PROTEIN MJ0541, MAGNESIUM ION | | Authors: | D'Angelo, I, Raffaelli, N, Dabusti, V, Lorenzi, T, Magni, G, Rizzi, M. | | Deposit date: | 2000-07-09 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of nicotinamide mononucleotide adenylyltransferase: a key enzyme in NAD(+) biosynthesis.
Structure Fold.Des., 8, 2000
|
|
4FCI
 
 | |
4FCK
 
 | |
4GWC
 
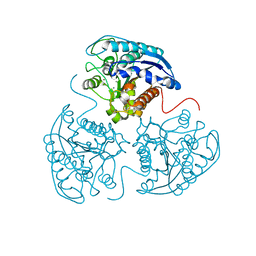 | | Crystal Structure of Mn2+2,Zn2+-Human Arginase I | | Descriptor: | Arginase-1, MANGANESE (II) ION, ZINC ION | | Authors: | D'Antonio, E.L, Hai, Y, Christianson, D.W. | | Deposit date: | 2012-09-01 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of non-native metal clusters in human arginase I.
Biochemistry, 51, 2012
|
|
4GSM
 
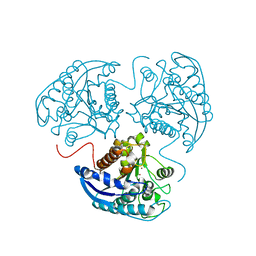 | |
4GWD
 
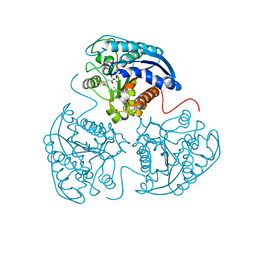 | | Crystal Structure of the Mn2+2,Zn2+-Human Arginase I-ABH Complex | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase-1, MANGANESE (II) ION, ... | | Authors: | D'Antonio, E.L, Hai, Y, Christianson, D.W. | | Deposit date: | 2012-09-01 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure and function of non-native metal clusters in human arginase I.
Biochemistry, 51, 2012
|
|
4GSZ
 
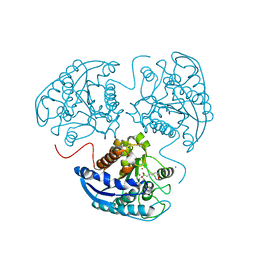 | |
4GSV
 
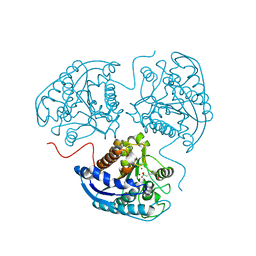 | |
3DRN
 
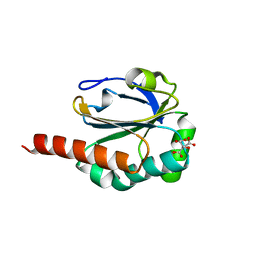 | |
7KUC
 
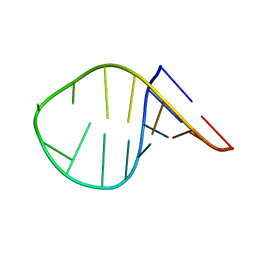 | |
7KUB
 
 | |
7KUD
 
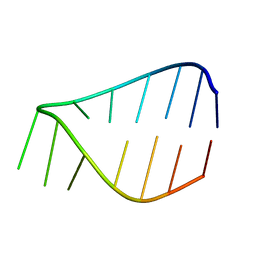 | |
2O00
 
 | | NMR structure analysis of the Penetratin conjugated Gas (374-394) peptide | | Descriptor: | Penetratin conjugated Gas (374-394) peptide | | Authors: | D'Ursi, A.M, Rovero, P, Albrizio, S, Espsito, C, D'Errico, G, Novellino, E. | | Deposit date: | 2006-11-27 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Driving forces in the delivery of penetratin conjugated G protein fragment.
J.Med.Chem., 50, 2007
|
|
2NZZ
 
 | | NMR structure analysis of the Penetratin conjugated Gas (374-394) peptide | | Descriptor: | Penetratin conjugated Gas (374-394) peptide | | Authors: | D'Ursi, A.M, Rovero, P, Albrizio, S, Espsito, C, D'Errico, G, Novellino, E. | | Deposit date: | 2006-11-27 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Driving forces in the delivery of penetratin conjugated G protein fragment.
J.Med.Chem., 50, 2007
|
|
2VGO
 
 | | Crystal structure of Aurora B kinase in complex with Reversine inhibitor | | Descriptor: | INNER CENTROMERE PROTEIN A, N~6~-cyclohexyl-N~2~-(4-morpholin-4-ylphenyl)-9H-purine-2,6-diamine, SERINE/THREONINE-PROTEIN KINASE 12-A | | Authors: | D'Alise, A.M, Amabile, G, Iovino, M, Di Giorgio, F.P, Bartiromo, M, Sessa, F, Villa, F, Musacchio, A, Cortese, R. | | Deposit date: | 2007-11-15 | | Release date: | 2008-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reversine, a Novel Aurora Kinases Inhibitor, Inhibits Colony Formation of Human Acute Myeloid Leukemia Cells.
Mol.Cancer Ther., 7, 2008
|
|
1S9S
 
 | |
4MNN
 
 | |
1QWP
 
 | | NMR analysis of 25-35 fragment of beta amyloid peptide | | Descriptor: | 11-mer peptide from Amyloid beta A4 protein | | Authors: | D'Ursi, A.M, Armenante, M.R, Guerrini, R, Salvadori, S, Sorrentino, G, Picone, D. | | Deposit date: | 2003-09-03 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide (25-35) in different media
J.Med.Chem., 47, 2004
|
|
1QYT
 
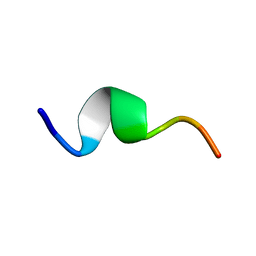 | | Solution structure of fragment (25-35) of beta amyloid peptide in SDS micellar solution | | Descriptor: | 11-mer peptide from Amyloid beta A4 protein | | Authors: | D'Ursi, A.M, Armenante, M.R, Guerrini, R, Salvadori, S, Sorrentino, G, Picone, D. | | Deposit date: | 2003-09-12 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide (25-35) in different media
J.Med.Chem., 47, 2004
|
|
2QP6
 
 | |
