7ONA
 
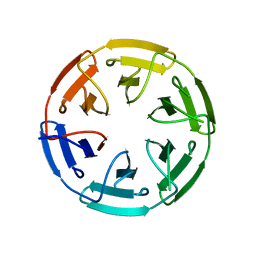 | | Crystal structure of the computationally designed SAKe6AC protein | | Descriptor: | CALCIUM ION, SAKe6AC | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
1LUN
 
 | | NMR Structure of the Itk SH2 domain, Pro287trans, energy minimized average structure | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Mallis, R.J, Brazin, K.N, Fulton, D.B, Andreotti, A.H. | | Deposit date: | 2002-05-22 | | Release date: | 2002-11-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of a proline-driven conformational switch
within the Itk SH2 domain
Nat.Struct.Biol., 9, 2002
|
|
7ONH
 
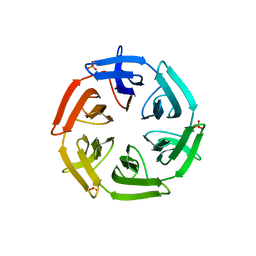 | | Crystal structure of the computationally designed SAKe6BE-L3 protein | | Descriptor: | SAKe6BE-L3, SULFATE ION | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
1M4E
 
 | | Solution Structure of Hepcidin-20 | | Descriptor: | Hepcidin | | Authors: | Hunter, H.N, Fulton, D.B, Ganz, T, Vogel, H.J. | | Deposit date: | 2002-07-02 | | Release date: | 2002-11-06 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human hepcidin, a peptide hormone with antimicrobial activity that is involved in iron uptake and hereditary hemochromatosis.
J.Biol.Chem., 277, 2002
|
|
1M4P
 
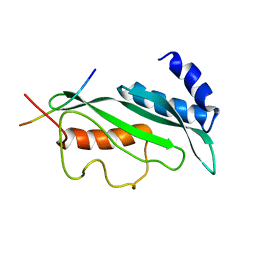 | | Structure of the Tsg101 UEV domain in complex with a HIV-1 PTAP "late domain" peptide, DYANA Ensemble | | Descriptor: | Gag Polyprotein, Tumor Susceptibility gene 101 protein | | Authors: | Pornillos, O, Alam, S.L, Davis, D.R, Sundquist, W.I. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein
Nat.Struct.Biol., 9, 2002
|
|
2VDA
 
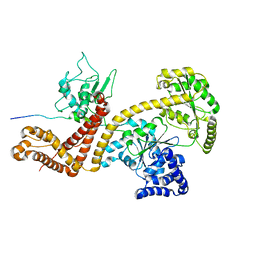 | | Solution structure of the SecA-signal peptide complex | | Descriptor: | MALTOPORIN, TRANSLOCASE SUBUNIT SECA | | Authors: | Gelis, I, Bonvin, A.M.J.J, Keramisanou, D, Koukaki, M, Gouridis, G, Karamanou, S, Economou, A, Kalodimos, C.G. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Signal-Sequence Recognition by the Translocase Motor Seca as Determined by NMR
Cell(Cambridge,Mass.), 131, 2007
|
|
1KLC
 
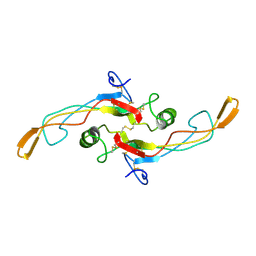 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
8GOP
 
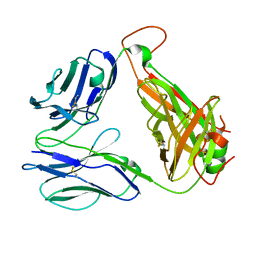 | | SARS-CoV-2 specific private TCR RLQ7 | | Descriptor: | SARS-CoV-2 specific private TCR RLQ7 alpha, SARS-CoV-2 specific private TCR RLQ7 beta | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into protection against a SARS-CoV-2 spike variant by T cell receptor (TCR) diversity.
J.Biol.Chem., 299, 2023
|
|
8GOM
 
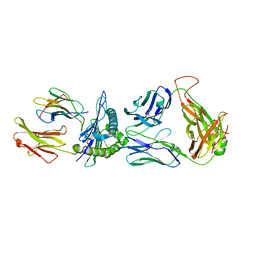 | | SARS-CoV-2 specific private TCR RLQ7 in complex with RLQ-HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, SARS-CoV-2 specific private TCR RLQ7 alpha, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.783 Å) | | Cite: | Structural insights into protection against a SARS-CoV-2 spike variant by T cell receptor (TCR) diversity.
J.Biol.Chem., 299, 2023
|
|
1KLD
 
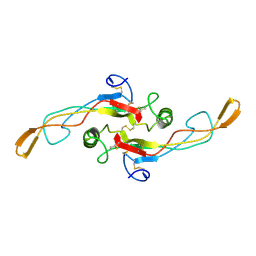 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MODELS 18-33 OF 33 STRUCTURES | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
8GON
 
 | |
1KSQ
 
 | | NMR Study of the Third TB Domain from Latent Transforming Growth Factor-beta Binding Protein-1 | | Descriptor: | LATENT TRANSFORMING GROWTH FACTOR BETA BINDING PROTEIN 1 | | Authors: | Lack, J, O'leary, J.M, Knott, V, Yuan, X, Rifkin, D.B, Handford, P.A, Downing, A.K. | | Deposit date: | 2002-01-14 | | Release date: | 2003-08-26 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Third TB Domain from LTBP1 Provides Insight into Assembly
of the Large Latent Complex that Sequesters Latent TGF-beta.
J.Mol.Biol., 334, 2003
|
|
1KZT
 
 | |
1L6U
 
 | | NMR STRUCTURE OF OXIDIZED ADRENODOXIN | | Descriptor: | Adrenodoxin 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Beilke, D, Weiss, R, Lohr, F, Pristovsek, P, Hannemann, F, Bernhardt, R, Rueterjans, H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A new electron transport mechanism in mitochondrial steroid hydroxylase systems based on structural changes upon the reduction of adrenodoxin.
Biochemistry, 41, 2002
|
|
1KZS
 
 | |
6L7K
 
 | | solution structure of hIFABP V60C/Y70C variant. | | Descriptor: | Fatty acid-binding protein, intestinal | | Authors: | Fan, J, Yang, D. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-04 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Ligand Entry into Fatty Acid Binding Protein via Local Unfolding Instead of Gap Widening.
Biophys.J., 118, 2020
|
|
1LIQ
 
 | | Non-native Solution Structure of a fragment of the CH1 domain of CBP | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Matthews, J.M, Kwan, A.H.Y, Newton, A, Gell, D.A, Crossley, M, Mackay, J.P. | | Deposit date: | 2002-04-18 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A New Zinc Binding Fold Underlines the Versatility of Zinc Binding Modules in Protein Evolution
Structure, 10, 2002
|
|
5NR6
 
 | |
8G7K
 
 | | mtHsp60 V72I apo focus | | Descriptor: | 60 kDa heat shock protein, mitochondrial | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7J
 
 | | mtHsp60 V72I apo | | Descriptor: | 60 kDa heat shock protein, mitochondrial | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
1L4S
 
 | |
6I6Y
 
 | |
7PSO
 
 | |
7PC6
 
 | | DNA-binding domain of a p53 homolog from the hydrothermal vent annelid Alvinella pompejana | | Descriptor: | 1,2-ETHANEDIOL, DNA-binding domain, ZINC ION | | Authors: | Balourdas, D.-I, Knapp, S, Soussi, T, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Evolutionary history of the p53 family DNA-binding domain: insights from an Alvinella pompejana homolog.
Cell Death Dis, 13, 2022
|
|
1LM0
 
 | | Solution structure and characterization of the heme chaperone CcmE | | Descriptor: | cytochrome c maturation protein E | | Authors: | Arnesano, F, Banci, L, Barker, P.D, Bertini, I, Rosato, A, Su, X.C, Viezzoli, M.S. | | Deposit date: | 2002-04-30 | | Release date: | 2002-12-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and characterization of the heme chaperone CcmE
Biochemistry, 41, 2002
|
|
