8JA1
 
 | |
4QA3
 
 | | Crystal structure of T311M HDAC8 in complex with Trichostatin A (TSA) | | Descriptor: | GLYCEROL, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Decroos, C, Bowman, C.B, Moser, J.-A.S, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2014-05-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.876 Å) | | Cite: | Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders.
Acs Chem.Biol., 9, 2014
|
|
2QV4
 
 | | Human pancreatic alpha-amylase complexed with nitrite and acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5R,6S)-4-{[alpha-D-glucopyranosyl-(1->4)-alpha-D-glucopyranosyl-(1->4)-alpha-D-glucopyranosyl]oxy}-5,6-dihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Williams, L.K, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-08-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
7L91
 
 | | Structure of Metallo Beta-Lactamase L1 in a Complex with Hydrolyzed Moxalactam Determined by Pink-Beam Serial Crystallography | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Vukica, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-01 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
4X1Y
 
 | |
6DVT
 
 | |
6P3O
 
 | | Tetrahydroprotoberberine N-methyltransferase in complex with (S)-cis-N-methylstylopine and S-adenosylhomocysteine | | Descriptor: | (5S,12bS)-5-methyl-6,7,12b,13-tetrahydro-2H,4H,10H-[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquinolino[3,2-a]isoquinolin-5-ium, S-ADENOSYL-L-HOMOCYSTEINE, Tetrahydroprotoberberine N-methyltransferase | | Authors: | Lang, D.E, Morris, J.S, Rowley, M, Torres, M.A, Maksimovich, V.A, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2019-05-24 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function studies of tetrahydroprotoberberineN-methyltransferase reveal the molecular basis of stereoselective substrate recognition.
J.Biol.Chem., 294, 2019
|
|
4X3H
 
 | | CRYSTAL STRUCTURE OF ARC N-LOBE COMPLEXED WITH STARGAZIN PEPTIDE | | Descriptor: | Activity-regulated cytoskeleton-associated protein, VOLTAGE-DEPENDENT CALCIUM CHANNEL GAMMA-2 SUBUNIT | | Authors: | zhang, W, ward, m, leahy, d, worley, p. | | Deposit date: | 2014-11-30 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis of arc binding to synaptic proteins: implications for cognitive disease.
Neuron, 86, 2015
|
|
4QNM
 
 | | CRYSTAL STRUCTURE of PSPF(1-265) E108Q MUTANT | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Psp operon transcriptional activator | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.628 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
8PRA
 
 | |
4X3I
 
 | | The crystal structure of Arc N-lobe complexed with CAMK2A fragment | | Descriptor: | Activity-regulated cytoskeleton-associated protein, CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE TYPE II SUBUNIT ALPHA | | Authors: | Zhang, W, Ward, M, Leahy, D, Worley, P. | | Deposit date: | 2014-11-30 | | Release date: | 2015-06-03 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of arc binding to synaptic proteins: implications for cognitive disease.
Neuron, 86, 2015
|
|
7A21
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2158 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(pyrrolidin-1-ylmethyl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type I, ... | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-08-14 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2153
To Be Published
|
|
8PRS
 
 | |
8JE0
 
 | | A novel amidohydrolase | | Descriptor: | 1,2-ETHANEDIOL, Amidase, ZINC ION | | Authors: | Ma, D, Feng, R. | | Deposit date: | 2023-05-15 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A novel amidohydrolase catalyze the degradation of PAM by Klebsiella sp. PCX
To Be Published
|
|
4QIH
 
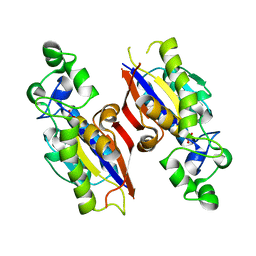 | | The structure of mycobacterial glucosyl-3-phosphoglycerate phosphatase Rv2419c complexes with VO3 | | Descriptor: | Glucosyl-3-phosphoglycerate phosphatase, VANADATE ION | | Authors: | Zhou, W.H, Zheng, Q.Q, Jiang, D.Q, Zhang, W, Zhang, Q.Q, Jin, J, Li, X, Yang, H.T, Shaw, N, Rao, Z. | | Deposit date: | 2014-05-30 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Mechanism of dephosphorylation of glucosyl-3-phosphoglycerate by a histidine phosphatase
J.Biol.Chem., 289, 2014
|
|
8FLI
 
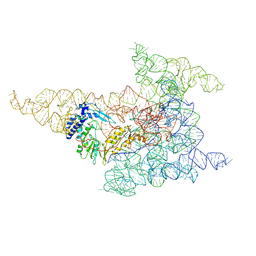 | | Cryo-EM structure of a group II intron immediately before branching | | Descriptor: | Group II Intron, MAGNESIUM ION, Maturase reverse transcriptase | | Authors: | Haack, D.B, Rudolfs, B.G, Zhang, C, Lyumkis, D, Toor, N. | | Deposit date: | 2022-12-21 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of branching during RNA splicing.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6P3M
 
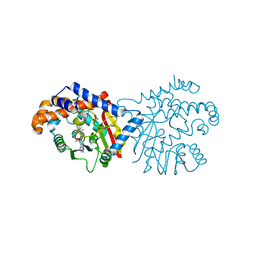 | | Tetrahydroprotoberberine N-methyltransferase in complex with S-adenosylhomocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, tetrahydroprotoberberine N-methyltransferase | | Authors: | Lang, D.E, Morris, J.S, Rowley, M, Torres, M.A, Maksimovich, V.A, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2019-05-24 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function studies of tetrahydroprotoberberineN-methyltransferase reveal the molecular basis of stereoselective substrate recognition.
J.Biol.Chem., 294, 2019
|
|
8DZ0
 
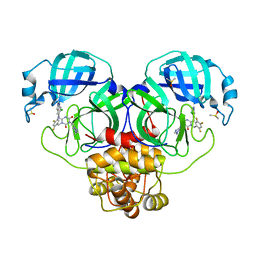 | | Crystal Structure of SARS-CoV-2 Main protease in complex with Ensitrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Oliva, G, Godoy, A.S. | | Deposit date: | 2022-08-06 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
6ZY1
 
 | |
7A96
 
 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound and 1 RBD Erect in Anticlockwise Direction | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
8DZ1
 
 | | Crystal Structure of SARS-CoV-2 Main protease mutant M49I in complex with Ensitrelvir | | Descriptor: | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab | | Authors: | Noske, G.D, Oliva, G, Godoy, A.S. | | Deposit date: | 2022-08-06 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
6DZV
 
 | | Wild type human serotonin transporter in complex with 15B8 Fab bound to ibogaine in occluded conformation | | Descriptor: | (5beta)-12-methoxyibogamine, 15B8 antibody heavy chain, 15B8 antibody light chain, ... | | Authors: | Coleman, J.A, Yang, D, Gouaux, E. | | Deposit date: | 2018-07-05 | | Release date: | 2019-04-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Serotonin transporter-ibogaine complexes illuminate mechanisms of inhibition and transport.
Nature, 569, 2019
|
|
5LLJ
 
 | |
5F6V
 
 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 1 (biphenol from fragment cocktail screen) | | Descriptor: | 2-(2-hydroxyphenyl)phenol, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
7QW7
 
 | | Adenine-specific DNA methyltransferase M.BseCI complexed with AdoHcy and cognate fully methylated DNA duplex | | Descriptor: | Fully methylated DNA duplex, Modification methylase BseCI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Mitsikas, D.A, Kouyianou, K, Kotsifaki, D, Providaki, M, Bouriotis, V, Glykos, N.M, Kokkinidis, M. | | Deposit date: | 2022-01-24 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of M.BseCI DNA methyltransferase from Geobacillus stearothermophilus.
To Be Published
|
|
