2VWR
 
 | | Crystal structure of the second pdz domain of numb-binding protein 2 | | Descriptor: | LIGAND OF NUMB PROTEIN X 2 | | Authors: | Roos, A.K, Guo, K, Burgess-Brown, N, Yue, W.W, Elkins, J.M, Pike, A.C.W, Filippakopoulos, P, Arrowsmith, C.H, Wikstom, M, Edwards, A, von Delft, F, Bountra, C, Doyle, D, Oppermann, U. | | Deposit date: | 2008-06-26 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of the Second Pdz Domain of the Human Numb-Binding Protein 2
To be Published
|
|
1XW2
 
 | | Structure Of A Cold-Adapted Family 8 Xylanase | | Descriptor: | Endo-1,4-beta-Xylanase | | Authors: | Collins, T, De Vos, D, Hoyoux, A, Savvides, S.N, Gerday, C, Van Beeumen, J, Feller, G. | | Deposit date: | 2004-10-29 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Study of the active site residues of a glycoside hydrolase family 8 xylanase
J.Mol.Biol., 354, 2005
|
|
1XX3
 
 | | Solution Structure of Escherichia coli TonB-CTD | | Descriptor: | TonB protein | | Authors: | Peacock, R.S, Weljie, A.M, Howard, S.P, Price, F.D, Vogel, H.J. | | Deposit date: | 2004-11-03 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the C-terminal domain of TonB and interaction studies with TonB box peptides
J.Mol.Biol., 345, 2005
|
|
5Q0X
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 6-(4-{[3-(3,5-dichloropyridin-4-yl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}-2-methylphenyl)-1-methyl-1H-indole-3-carbox ylic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
2QHV
 
 | | Structural Basis of Octanoic Acid Recognition by Lipoate-Protein Ligase B | | Descriptor: | Lipoyltransferase, OCTAN-1-OL | | Authors: | Kim, D.J, Lee, S.J, Kim, H.S, Kim, K.H, Lee, H.H, Yoon, H.J, Suh, S.W. | | Deposit date: | 2007-07-03 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of octanoic acid recognition by lipoate-protein ligase B
Proteins, 70, 2008
|
|
2XCZ
 
 | | Crystal Structure of macrophage migration inhibitory factor homologue from Prochlorococcus marinus | | Descriptor: | DI(HYDROXYETHYL)ETHER, POSSIBLE ATLS1-LIKE LIGHT-INDUCIBLE PROTEIN | | Authors: | Wasiel, A.A, Rozeboom, H.J, Hauke, D, Baas, B.J, Zandvoort, E, Quax, W.J, Thunnissen, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2010-04-27 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural and Functional Characterization of a Macrophage Migration Inhibitory Factor Homologue from the Marine Cyanobacterium Prochlorococcus Marinus.
Biochemistry, 49, 2010
|
|
2QHT
 
 | | Structural Basis of Octanoic Acid Recognition by Lipoate-Protein Ligase B | | Descriptor: | Lipoyltransferase | | Authors: | Kim, D.J, Lee, S.J, Kim, H.S, Kim, K.H, Lee, H.H, Yoon, H.J, Suh, S.W. | | Deposit date: | 2007-07-02 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of octanoic acid recognition by lipoate-protein ligase B
Proteins, 70, 2008
|
|
5Q0T
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 2-phenyl-N-(propan-2-yl)-6-[(thiophen-2-yl)sulfonyl]-4,5,6,7-tetrahydro-1H-pyrrolo[2,3-c]pyridine-1-carboxamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
1MV1
 
 | | The Tandem, Sheared PA Pairs in 5'(rGGCPAGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*(P5P)P*AP*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition in Purine-Rich Internal Loops: Thermodynamic, Structural, and Dynamic Consequences of Purine for Adenine Substitutions in 5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
5Q11
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N,N-dicyclohexyl-3-(2,4-dichlorophenyl)-5-methyl-1,2-oxazole-4-carboxamide | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
2FHT
 
 | | Crystal Structure of Viral Macrophage Inflammatory Protein-II | | Descriptor: | Viral macrophage inflammatory protein-II | | Authors: | Li, Y, Liu, D, Cao, R, Kumar, S, Dong, C.Z, wilson, S.R, Gao, Y.G, Huang, Z. | | Deposit date: | 2005-12-27 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of chemically synthesized vMIP-II.
Proteins, 67, 2007
|
|
1Y4U
 
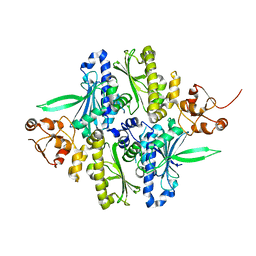 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | Chaperone protein htpG | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
2ETZ
 
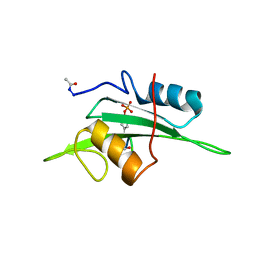 | | The NMR minimized average structure of the Itk SH2 domain bound to a phosphopeptide | | Descriptor: | Lymphocyte cytosolic protein 2 phosphopeptide fragment, Tyrosine-protein kinase ITK/TSK | | Authors: | Sundd, M, Pletneva, E.V, Fulton, D.B, Andreotti, A.H. | | Deposit date: | 2005-10-27 | | Release date: | 2006-02-07 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Molecular Details of Itk Activation by Prolyl Isomerization and Phospholigand Binding: The NMR Structure of the Itk SH2 Domain Bound to a Phosphopeptide.
J.Mol.Biol., 357, 2006
|
|
5OU3
 
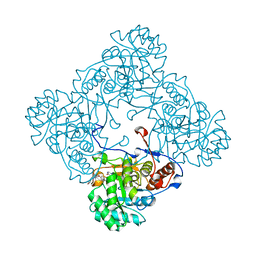 | | M. thermoresistible IMPDH in complex with IMP and Compound 31 (AT080) | | Descriptor: | (2~{S})-~{N}-[5-(4-bromophenyl)-1~{H}-imidazol-2-yl]-2-[4-(1-methylimidazol-4-yl)phenoxy]propanamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2017-08-23 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-Based Approach to Targeting Inosine-5'-monophosphate Dehydrogenase (IMPDH) from Mycobacterium tuberculosis.
J. Med. Chem., 61, 2018
|
|
2AQC
 
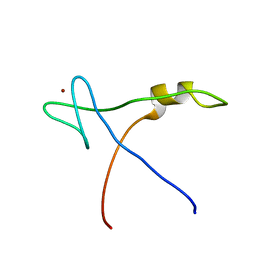 | | NMR Structural analysis of archaeal Nop10 | | Descriptor: | Ribosome biogenesis protein Nop10, ZINC ION | | Authors: | Hamma, T, Reichow, S.L, Varani, G, Ferre-D'Amare, A.R. | | Deposit date: | 2005-08-17 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Cbf5-Nop10 complex is a molecular bracket that organizes box H/ACA RNPs.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2XWS
 
 | | ANAEROBIC COBALT CHELATASE (CbiX) FROM ARCHAEOGLOBUS FULGIDUS | | Descriptor: | SIROHYDROCHLORIN COBALTOCHELATASE | | Authors: | Romao, C.V, Ladakis, D, Lobo, S.A.L, Carrondo, M.A, Brindley, A.A, Deery, E, Matias, P.M, Pickersgill, R.W, Saraiva, L.M, Warren, M.J. | | Deposit date: | 2010-11-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution in a Family of Chelatases Facilitated by the Introduction of Active Site Asymmetry and Protein Oligomerization.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4BK4
 
 | | crystal structure of the human EphA4 ectodomain | | Descriptor: | EPHRIN TYPE-A RECEPTOR 4 | | Authors: | Seiradake, E, Schaupp, A, del Toro Ruiz, D, Kaufmann, R, Mitakidis, N, Harlos, K, Aricescu, A.R, Klein, R, Jones, E.Y. | | Deposit date: | 2013-04-22 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structurally Encoded Intraclass Differences in Epha Clusters Drive Distinct Cell Responses
Nat.Struct.Mol.Biol., 20, 2013
|
|
2AZS
 
 | | NMR structure of the N-terminal SH3 domain of Drk (calculated without NOE restraints) | | Descriptor: | SH2-SH3 adapter protein drk | | Authors: | Bezsonova, I, Singer, A.U, Choy, W.-Y, Tollinger, M, Forman-Kay, J.D. | | Deposit date: | 2005-09-12 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Comparison of the Unstable drkN SH3 Domain and a Stable Mutant
Biochemistry, 44, 2005
|
|
2CRS
 
 | |
1M62
 
 | | Solution structure of the BAG domain from BAG4/SODD | | Descriptor: | BAG-family molecular chaperone regulator-4 | | Authors: | Briknarova, K, Takayama, S, Homma, S, Baker, K, Cabezas, E, Hoyt, D.W, Li, Z, Satterthwait, A.C, Ely, K.R. | | Deposit date: | 2002-07-11 | | Release date: | 2002-07-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | BAG4/SODD protein contains a short BAG domain.
J.Biol.Chem., 277, 2002
|
|
2P5H
 
 | | sPLA2 inhibitor 9 | | Descriptor: | pip9 | | Authors: | Thwin, M.M, Satyanarayanajois, D.S, Nagarajarao, L.M, Sato, K, Gopalakrishnakone, P.P, Arjunan, P. | | Deposit date: | 2007-03-15 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Novel Peptide Inhibitors of Human Secretory Phospholipase A2 with Antiinflammatory Activity: Solution Structure and Molecular Modeling.
J.Med.Chem., 50, 2007
|
|
2OUQ
 
 | | crystal structure of PDE10A2 in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5R5Z
 
 | | PanDDA analysis group deposition -- Crystal Structure of FIBRINOGEN-LIKE GLOBE DOMAIN OF HUMAN TENASCIN-C in complex with Z2856434821 | | Descriptor: | 2-methyl-2-{[(3-methylthiophen-2-yl)methyl]amino}propan-1-ol, Tenascin C (Hexabrachion), isoform CRA_a | | Authors: | Coker, J.A, Bezerra, G.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Yue, W.W, Marsden, B.D. | | Deposit date: | 2020-02-28 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2A8Z
 
 | | Structure Of A Cold-Adapted Family 8 Xylanase | | Descriptor: | endo-1,4-beta-xylanase | | Authors: | Collins, T, De Vos, D, Hoyoux, A, Savvides, S.N, Gerday, C, Van Beeumen, J, Feller, G. | | Deposit date: | 2005-07-10 | | Release date: | 2005-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Study of the active site residues of a glycoside hydrolase family 8 xylanase.
J.Mol.Biol., 354, 2005
|
|
5R5T
 
 | | PanDDA analysis group deposition -- Crystal Structure of FIBRINOGEN-LIKE GLOBE DOMAIN OF HUMAN TENASCIN-C in complex with Z1251207602 | | Descriptor: | 1-(5-methyl-1,3,4-thiadiazol-2-yl)piperidine, Tenascin C (Hexabrachion), isoform CRA_a | | Authors: | Coker, J.A, Bezerra, G.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Yue, W.W, Marsden, B.D. | | Deposit date: | 2020-02-28 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
