2QOA
 
 | |
2ESD
 
 | | Crystal Structure of thioacylenzyme intermediate of an Nadp Dependent Aldehyde Dehydrogenase | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase | | Authors: | D'Ambrosio, K, Didierjean, C, Benedetti, E, Aubry, A, Corbier, C. | | Deposit date: | 2005-10-26 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The first crystal structure of a thioacylenzyme intermediate in the ALDH family: new coenzyme conformation and relevance to catalysis
Biochemistry, 45, 2006
|
|
2D4Q
 
 | | Crystal structure of the Sec-PH domain of the human neurofibromatosis type 1 protein | | Descriptor: | Neurofibromin, OXTOXYNOL-10, PYROPHOSPHATE 2- | | Authors: | D'angelo, I, Welti, S, Bonneau, F, Scheffzek, K. | | Deposit date: | 2005-10-22 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel bipartite phospholipid-binding module in the neurofibromatosis type 1 protein
Embo Rep., 7, 2006
|
|
1QXC
 
 | | NMR structure of the fragment 25-35 of beta amyloid peptide in 20/80 v:v hexafluoroisopropanol/water mixture | | Descriptor: | 11-mer peptide from Amyloid beta A4 protein | | Authors: | D'Ursi, A.M, Armenante, M.R, Guerrini, R, Salvadori, S, Sorrentino, G, Picone, D. | | Deposit date: | 2003-09-05 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide (25-35) in different media
J.Med.Chem., 47, 2004
|
|
3O27
 
 | |
3V5G
 
 | |
2E2X
 
 | |
2HLS
 
 | |
2MS0
 
 | |
1U6P
 
 | |
2MS1
 
 | |
2MQT
 
 | |
2JSI
 
 | | 11-23 obestatin fragment in DPC/SDS micellar solution | | Descriptor: | Appetite-regulating hormone, Obestatin | | Authors: | D'Ursi, A.M, Scrima, M, Esposito, C, Campiglia, P. | | Deposit date: | 2007-07-05 | | Release date: | 2008-10-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Obestatin conformational features: a strategy to unveil obestatin's biological role?
Biochem.Biophys.Res.Commun., 363, 2007
|
|
2JSH
 
 | | obestatin NMR structure in SDS/DPC micellar solution | | Descriptor: | Appetite-regulating hormone, Obestatin | | Authors: | D'Ursi, A.M, Scrima, M, Esposito, C, Campiglia, P. | | Deposit date: | 2007-07-05 | | Release date: | 2008-10-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Obestatin conformational features: a strategy to unveil obestatin's biological role?
Biochem.Biophys.Res.Commun., 363, 2007
|
|
2JSJ
 
 | | Obestatin in water solution | | Descriptor: | Appetite-regulating hormone, Obestatin | | Authors: | D'Ursi, A.M, Scrima, M, Esposito, C, Campiglia, P. | | Deposit date: | 2007-07-05 | | Release date: | 2008-10-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Obestatin conformational features: a strategy to unveil obestatin's biological role?
Biochem.Biophys.Res.Commun., 363, 2007
|
|
2MQV
 
 | |
4UQH
 
 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the inhibitor (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(3,4-difluorophenyl)piperazin-1-yl)-2-fluorobenzamide. | | Descriptor: | (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(3,4-difluorophenyl)piperazin-1-yl)-2-fluorobenzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE, ... | | Authors: | Calvet, C.M, Vieira, D.F, Choi, J.Y, Cameron, M.D, Gut, J, Kellar, D, Siqueira-Neto, J.L, McKerrow, J.H, Roush, W.R, Podust, L.M. | | Deposit date: | 2014-06-23 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | 4-Aminopyridyl-Based Cyp51 Inhibitors as Anti-Trypanosoma Cruzi Drug Leads with Improved Pharmacokinetic Profile and in Vivo Potency.
J.Med.Chem., 57, 2014
|
|
4WCU
 
 | | PDE4 complexed with inhibitor | | Descriptor: | MAGNESIUM ION, N-benzyl-2-{6-[(3,5-dichloropyridin-4-yl)acetyl]-2,3-dimethoxyphenoxy}acetamide, ZINC ION, ... | | Authors: | Sorensen, M.D. | | Deposit date: | 2014-09-05 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery and Early Clinical Development of 2-{6-[2-(3,5-Dichloro-4-pyridyl)acetyl]-2,3-dimethoxyphenoxy}-N-propylacetamide (LEO 29102), a Soft-Drug Inhibitor of Phosphodiesterase 4 for Topical Treatment of Atopic Dermatitis.
J. Med. Chem., 57, 2014
|
|
4V25
 
 | | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context- dependent cytostasis in cancer cells | | Descriptor: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, N-[4-(2-chloro-5-methylpyrimidin-4-yl)phenyl]-N-(4-{[(difluoroacetyl)amino]methyl}benzyl)-2,4-dihydroxybenzamide, ... | | Authors: | Moore, J.D, Staniszewska, A, Shaw, T, D'Alessandro, J, Davis, B, Surgenor, A, Baker, L, Matassova, N, Murray, J, Macias, A, Brough, P, Wood, M, Mahon, P.C. | | Deposit date: | 2014-10-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context-dependent cytostasis in cancer cells.
Oncotarget, 5, 2014
|
|
4W8A
 
 | | Crystal structure of XEG5B, a GH5 xyloglucan-specific beta-1,4-glucanase from ruminal metagenomic library, in the native form | | Descriptor: | Exo-xyloglucanase, GLYCEROL, SULFATE ION | | Authors: | Santos, C.R, Cordeiro, R.L, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for Xyloglucan Specificity and alpha-d-Xylp(1 6)-d-Glcp Recognition at the -1 Subsite within the GH5 Family.
Biochemistry, 54, 2015
|
|
4V4C
 
 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ACETATE ION, CALCIUM ION, ... | | Authors: | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | Deposit date: | 2004-06-02 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
4V4E
 
 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici complexed with inhibitor 1,2,4,5-tetrahydroxy-benzene | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, BENZENE-1,2,4,5-TETROL, CALCIUM ION, ... | | Authors: | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | Deposit date: | 2004-06-02 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
4UT3
 
 | | X-ray structure of the human PP1 gamma catalytic subunit treated with hydrogen peroxide | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, SERINE/THREONINE-PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT | | Authors: | Zeh Silva, M, Kopec, J, Fotinou, D, Steiner, R.A. | | Deposit date: | 2014-07-17 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Targeted Redox Inhibition of Protein Phosphatase 1 by Nox4 Regulates Eif2Alpha-Mediated Stress Signaling.
Embo J., 35, 2016
|
|
4UV4
 
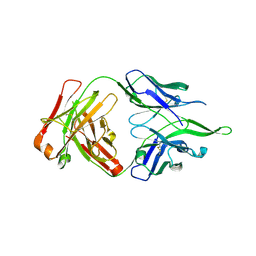 | | Crystal structure of anti-FPR Fpro0165 Fab fragment | | Descriptor: | FPRO0165 FAB | | Authors: | Douthwaite, J.A, Sridharan, S, Huntington, C, Marwood, R, Hammersley, J, Hakulinen, J.K, Ek, M, Sjogren, T, Rider, D, Privezentzev, C, Seaman, J.C, Cariuk, P, Knights, V, Young, J, Wilkinson, T, Sleeman, M, Finch, D.K, Lowe, D.C, Vaughan, T.J. | | Deposit date: | 2014-08-04 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Affinity Maturation of a Novel Antagonistic Human Monoclonal Antibody with a Long Vh Cdr3 Targeting the Class a Gpcr Formyl-Peptide Receptor 1.
Mabs, 7, 2015
|
|
4UUP
 
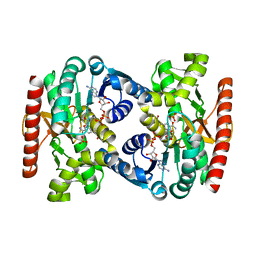 | | Reconstructed ancestral trichomonad malate dehydrogenase in complex with NADH, SO4, and PO4 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MALATE DEHYDROGENASE, PHOSPHATE ION, ... | | Authors: | Steindel, P.A, Chen, E.H, Theobald, D.L. | | Deposit date: | 2014-07-29 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Gradual Neofunctionalization in the Convergent Evolution of Trichomonad Lactate and Malate Dehydrogenases.
Protein Sci., 25, 2016
|
|
