3GMV
 
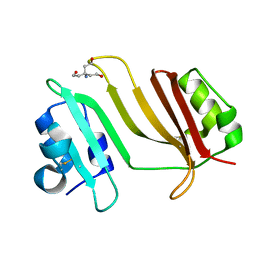 | | Crystal Structure of Beta-Lactamse Inhibitory Protein-I (BLIP-I) in Apo Form | | Descriptor: | Beta-lactamase inhibitory protein BLIP-I, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Lim, D.C, Gretes, M, Strynadka, N.C.J. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
7RV6
 
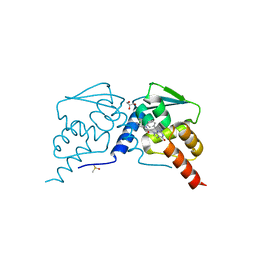 | |
1L6H
 
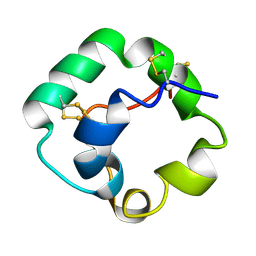 | |
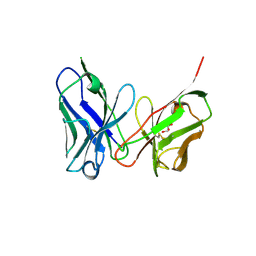
8QT4
 
 | | Crystal structure of human Sirt2 in complex with the super-slow substrate TNFn-6 and NAD+ | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Friedrich, F, Kalbas, D, Meleshin, M, Einsle, O, Schutkowski, M, Jung, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | New Super-Slow Substrates as novel Sirtuin-Inhibitors
To Be Published
|
|
8H3B
 
 | | Crystal structure of antibody scFv against M2e Influenza peptide | | Descriptor: | GLYCEROL, Single Chain Variable Fragment | | Authors: | Kumar, U, Madni, Z.K, Gaur, V, Salunke, D.M. | | Deposit date: | 2022-10-08 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A structure and knowledge-based combinatorial approach to engineering universal scFv antibodies against influenza M2 protein.
J.Biomed.Sci., 30, 2023
|
|
8QT3
 
 | | Crystal structure of human Sirt2 in complex with the super-slow substrate TNFn-5 and NAD+ | | Descriptor: | (2S)-2-dodecylsulfanylpropanoic acid, 1,2-ETHANEDIOL, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Friedrich, F, Kalbas, D, Meleshin, M, Einsle, O, Schutkowski, M, Jung, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | New Super-Slow Substrates as novel Sirtuin-Inhibitors
To Be Published
|
|
8QTU
 
 | | Crystal structure of human Sirt2 in complex with the super-slow substrate TNFn-3 and NAD+ | | Descriptor: | 1,2-ETHANEDIOL, 3-dodecylsulfanyl-3-methyl-butanoic acid, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Friedrich, F, Kalbas, D, Meleshin, M, Einsle, O, Schutkowski, M, Jung, M. | | Deposit date: | 2023-10-13 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New Super-Slow Substrates as novel Sirtuin-Inhibitors
To Be Published
|
|
6Q1D
 
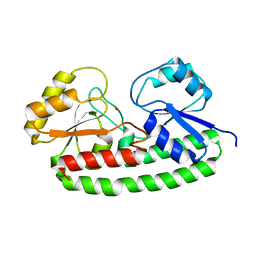 | | Holo YfeA reconstituted by zinc soaking | | Descriptor: | Periplasmic chelated iron-binding protein YfeA, ZINC ION | | Authors: | Radka, C.D, Labiuk, S.L, DeLucas, L.J, Aller, S.G. | | Deposit date: | 2019-08-03 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structures of the substrate-binding protein YfeA in apo and zinc-reconstituted holo forms.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7RV9
 
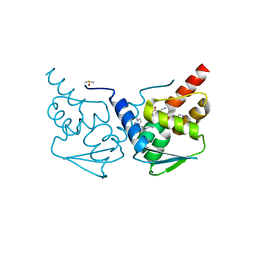 | | Crystal structure of the BCL6 BTB domain in complex with OICR-10269 | | Descriptor: | DIMETHYL SULFOXIDE, Isoform 2 of B-cell lymphoma 6 protein, N-[5-chloro-2-(4-methylpiperazin-1-yl)pyridin-4-yl]-2-{5-(3-cyano-4-hydroxy-5-methylphenyl)-3-[3-(1-methyl-1H-pyrazol-4-yl)prop-2-yn-1-yl]-4-oxo-3,4-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl}acetamide | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-08-18 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the BCL6 BTB domain
To Be Published
|
|
6Q7K
 
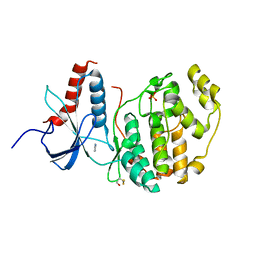 | | ERK2 mini-fragment binding | | Descriptor: | 1H-imidazol-2-amine, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
7PSO
 
 | |
8CEF
 
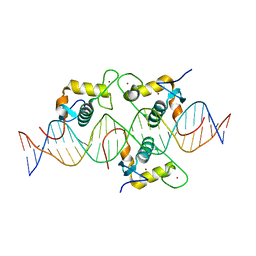 | | Asymmetric Dimerization in a Transcription Factor Superfamily is Promoted by Allosteric Interactions with DNA | | Descriptor: | DNA (26-MER), Nuclear receptor DNA binding domain, ZINC ION | | Authors: | Patel, A.K.M, Shaik, T.B, McEwen, A.G, Moras, D, Klaholz, B.P, Billas, I.M.L. | | Deposit date: | 2023-02-01 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Asymmetric dimerization in a transcription factor superfamily is promoted by allosteric interactions with DNA.
Nucleic Acids Res., 51, 2023
|
|
6T39
 
 | | Crystal structure of rsEGFP2 in its off-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Coquelle, N, Adam, V, Barends, T.R.M, De La Mora, E, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Photoswitching mechanism of a fluorescent protein revealed by time-resolved crystallography and transient absorption spectroscopy.
Nat Commun, 11, 2020
|
|
5E8K
 
 | | Crystal structure of polyprenyl pyrophosphate synthase 2 from Arabidopsis thaliana | | Descriptor: | Geranylgeranyl pyrophosphate synthase 10, mitochondrial | | Authors: | Wang, C, Chen, Q, Fan, D, Li, J, Wang, G, Zhang, P. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.028 Å) | | Cite: | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
8E04
 
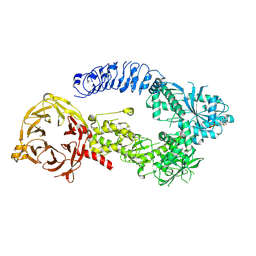 | | Structure of monomeric LRRK1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 1 | | Authors: | Reimer, J.M, Mathea, S, Chatterjee, D, Knapp, S, Leschziner, A.E. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of LRRK1 and mechanisms of autoinhibition and activation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7N17
 
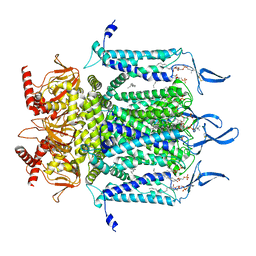 | | Structure of TAX-4_R421W apo open state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
7N15
 
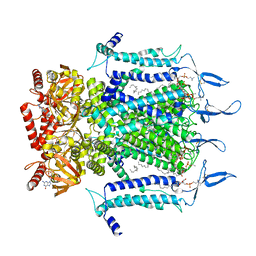 | | Structure of TAX-4_R421W w/cGMP open state | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CYCLIC GUANOSINE MONOPHOSPHATE, ... | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
7N16
 
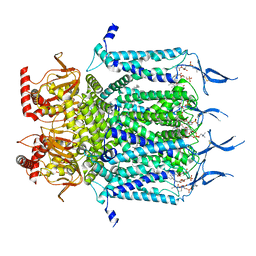 | | Structure of TAX-4_R421W apo closed state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, SODIUM ION | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
5EAM
 
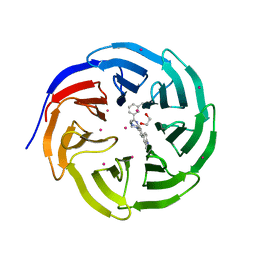 | | Crystal structure of human WDR5 in complex with compound 9o | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
8H2D
 
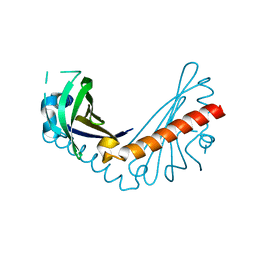 | |
7PC6
 
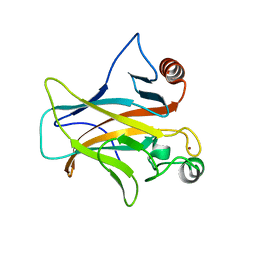 | | DNA-binding domain of a p53 homolog from the hydrothermal vent annelid Alvinella pompejana | | Descriptor: | 1,2-ETHANEDIOL, DNA-binding domain, ZINC ION | | Authors: | Balourdas, D.-I, Knapp, S, Soussi, T, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Evolutionary history of the p53 family DNA-binding domain: insights from an Alvinella pompejana homolog.
Cell Death Dis, 13, 2022
|
|
7NAK
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (TIR:1AD) | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(5-iodanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
5HPR
 
 | | Insulin with proline analog HyP at position B28 in the T2 state | | Descriptor: | GLYCEROL, Insulin A-Chain, Insulin B-Chain, ... | | Authors: | Lieblich, S.A, Fang, K.Y, Cahn, J.K.B, Tirrell, D.A. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | 4S-Hydroxylation of Insulin at ProB28 Accelerates Hexamer Dissociation and Delays Fibrillation.
J. Am. Chem. Soc., 139, 2017
|
|
7NAL
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (ARM and SAM domains) | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1 | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
6VQZ
 
 | | HLA-B*27:05 presenting an HIV-1 6mer peptide | | Descriptor: | 6-mer peptide, ARGININE, Beta-2-microglobulin, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
